| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,079,652 – 12,079,794 |
| Length | 142 |
| Max. P | 0.925625 |

| Location | 12,079,652 – 12,079,754 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 92.69 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -27.12 |
| Energy contribution | -28.62 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
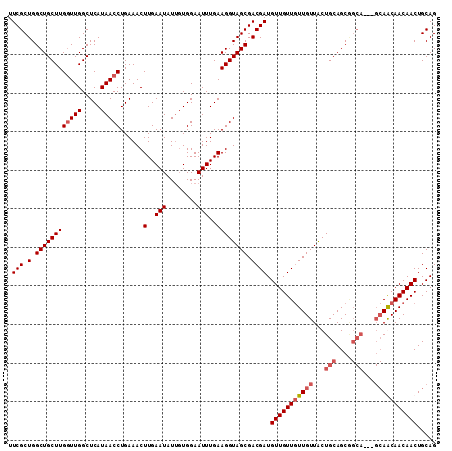
>3R_DroMel_CAF1 12079652 102 + 27905053 UUCGCUGGCUGCUUGGUUGGCUCAUAACCUGAAACUUGAAUAUUGUGGAAUUUGAAGGUAGCGACGAUGUUGUUUUUGUUACUGCAGC---------AGCAACAACUGCAG .(((.(.((((((((((((.....)))))......(..(((........)))..)))))))).))))(((((((.((((....)))).---------)))))))....... ( -25.90) >DroSec_CAF1 16391 108 + 1 UUCGCUGGCUGCUUGGUUGGCUCAUAACCUGAAACUUGAAUAUUGUGGAAUUUGAAGGUAGCGACGAUGUUGUUGUUGUUACUGCAGCGGCA---GCAACAACAACUGCAG .(((.(.((((((((((((.....)))))......(..(((........)))..)))))))).))))(((.(((((((((.((((....)))---).))))))))).))). ( -36.10) >DroSim_CAF1 16578 108 + 1 UUCGCUGGCUGCUUGGUUGGCUCAUAACCUGAAACUUGAAUAUUGUGGAAUUUGAAGGUAGCGACGAUGUUGUUGUUGUUACUGCAGCGGCA---GCAACAACAACUGCAG .(((.(.((((((((((((.....)))))......(..(((........)))..)))))))).))))(((.(((((((((.((((....)))---).))))))))).))). ( -36.10) >DroEre_CAF1 18425 111 + 1 UUCGCUGGCUGCUUGUUUGGCUCAUAACCUGAAACUUGAAUAUGUUGGAAUUUGAAGGUAGCGACGAUGUUGUUGUUGUUGCUGCAGCAGCAACAGCAGCAACAACUGCAG .((((((.((.(..((((.(..((((..(........)..)))).).))))..).)).))))))...(((((((((((((((((...)))))))))))))))))....... ( -39.60) >consensus UUCGCUGGCUGCUUGGUUGGCUCAUAACCUGAAACUUGAAUAUUGUGGAAUUUGAAGGUAGCGACGAUGUUGUUGUUGUUACUGCAGCGGCA___GCAACAACAACUGCAG .(((.(.((((((((((((.....)))))......(..(((........)))..)))))))).)))).(((((((((((...(((....)))...)))))))))))..... (-27.12 = -28.62 + 1.50)


| Location | 12,079,692 – 12,079,794 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 93.15 |
| Mean single sequence MFE | -39.75 |
| Consensus MFE | -32.95 |
| Energy contribution | -34.20 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
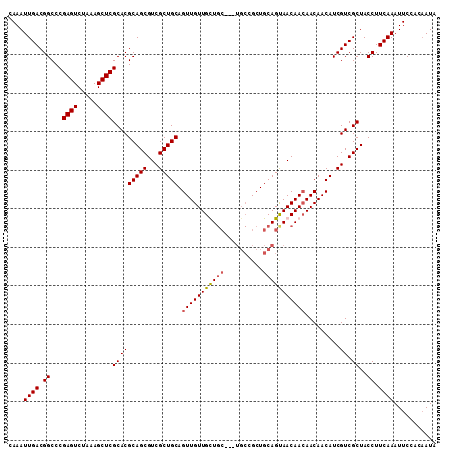
>3R_DroMel_CAF1 12079692 102 + 27905053 UAUUGUGGAAUUUGAAGGUAGCGACGAUGUUGUUUUUGUUACUGCAGC---------AGCAACAACUGCAGCGACGCUGCGUGCGAGCUUUAGACUCGGGCCGUCAAUUUG ......................((((.(((((((.((((....)))).---------)))))))...(((((...)))))((.(((((....).)))).))))))...... ( -29.30) >DroSec_CAF1 16431 108 + 1 UAUUGUGGAAUUUGAAGGUAGCGACGAUGUUGUUGUUGUUACUGCAGCGGCA---GCAACAACAACUGCAGCGACGCUGCGUGCGAGCUUUAGACUCGGGCCGUCAAUUUG ...........((((.(((((((.((.(((.(((((((((.((((....)))---).))))))))).))).)).))))))((.(((((....).)))).))).)))).... ( -41.70) >DroSim_CAF1 16618 108 + 1 UAUUGUGGAAUUUGAAGGUAGCGACGAUGUUGUUGUUGUUACUGCAGCGGCA---GCAACAACAACUGCAGCGACGCUGCGUGCGAGCUUUAGACUCGGGCCGUCAAUUUG ...........((((.(((((((.((.(((.(((((((((.((((....)))---).))))))))).))).)).))))))((.(((((....).)))).))).)))).... ( -41.70) >DroEre_CAF1 18465 111 + 1 UAUGUUGGAAUUUGAAGGUAGCGACGAUGUUGUUGUUGUUGCUGCAGCAGCAACAGCAGCAACAACUGCAGCGACGCUGCGUGCGAGCUUUAGACUCGGGCCGUCAAUUUG ...(((.((.(((((((.(.((.((..(((((((((((((((((...)))))))))))))))))...(((((...))))))))).).))))))).)).))).......... ( -46.30) >consensus UAUUGUGGAAUUUGAAGGUAGCGACGAUGUUGUUGUUGUUACUGCAGCGGCA___GCAACAACAACUGCAGCGACGCUGCGUGCGAGCUUUAGACUCGGGCCGUCAAUUUG ...........((((.(((((((.((..(((((((((((...(((....)))...))))))))))).....)).))))))((.(((((....).)))).))).)))).... (-32.95 = -34.20 + 1.25)



| Location | 12,079,692 – 12,079,794 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 93.15 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -28.51 |
| Energy contribution | -28.82 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.92 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12079692 102 - 27905053 CAAAUUGACGGCCCGAGUCUAAAGCUCGCACGCAGCGUCGCUGCAGUUGUUGCU---------GCUGCAGUAACAAAAACAACAUCGUCGCUACCUUCAAAUUCCACAAUA ......((((((.(((((.....)))))...))...((..(((((((.......---------)))))))..))...........))))...................... ( -26.80) >DroSec_CAF1 16431 108 - 1 CAAAUUGACGGCCCGAGUCUAAAGCUCGCACGCAGCGUCGCUGCAGUUGUUGUUGC---UGCCGCUGCAGUAACAACAACAACAUCGUCGCUACCUUCAAAUUCCACAAUA ....((((.((..(((((.....))))).....((((.((.((..(((((((((((---(((....))))))))))))))..)).)).)))).)).))))........... ( -40.40) >DroSim_CAF1 16618 108 - 1 CAAAUUGACGGCCCGAGUCUAAAGCUCGCACGCAGCGUCGCUGCAGUUGUUGUUGC---UGCCGCUGCAGUAACAACAACAACAUCGUCGCUACCUUCAAAUUCCACAAUA ....((((.((..(((((.....))))).....((((.((.((..(((((((((((---(((....))))))))))))))..)).)).)))).)).))))........... ( -40.40) >DroEre_CAF1 18465 111 - 1 CAAAUUGACGGCCCGAGUCUAAAGCUCGCACGCAGCGUCGCUGCAGUUGUUGCUGCUGUUGCUGCUGCAGCAACAACAACAACAUCGUCGCUACCUUCAAAUUCCAACAUA ....((((.((...((((.....))))(((((((((...))))).(((((((((((.((....)).))))))))))).........)).))..)).))))........... ( -38.80) >consensus CAAAUUGACGGCCCGAGUCUAAAGCUCGCACGCAGCGUCGCUGCAGUUGUUGCUGC___UGCCGCUGCAGUAACAACAACAACAUCGUCGCUACCUUCAAAUUCCACAAUA ....((((.((...((((.....))))(((((((((...))))).(((((((((((..........))))))))))).........)).))..)).))))........... (-28.51 = -28.82 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:40 2006