
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,077,607 – 12,077,824 |
| Length | 217 |
| Max. P | 0.989199 |

| Location | 12,077,607 – 12,077,719 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -27.96 |
| Energy contribution | -27.48 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12077607 112 + 27905053 GC---UAACUUCGCUGCCGGCGUCGCUGCUUAUAAAUCUUCGACAUGAAGC--GCAACAAAAAGCCGCAAAAAA---AGAAGGCAGAAACAGCAAGCGGCGCGCAUUGAAAAUUGCUCUA ((---....((((.(((..(((((((((((......(((((........((--((........).)))......---.))))).......))).))))))))))).))))....)).... ( -30.68) >DroSec_CAF1 14356 116 + 1 GCCGCCAACUUCGCUGCCGGCGUCGCUGCUUAUAAAUCUUCGACAUGAAGC--GCAACAAAAAGCCGCAAAAAA--AAGAAGGCAGAAACAGCAAGCGGCGCGCAUUGAAAAUUGCUCUA ...((....((((.(((..(((((((((((......(((((........((--((........).)))......--..))))).......))).))))))))))).))))....)).... ( -30.71) >DroSim_CAF1 14551 115 + 1 GC---CAACUUCGCUGCCGGCGUCGCUGCUUAUAAAUCUUCGACAUGAAGC--GCAACAAAAAGCCGCAAAAAAAGAAGAAGGCAGAAACAGCAAGCGGCGCGCAUUGAAAAUUGCUCUA ..---......((((((..((((..((((((.....(((((........((--((........).))).......)))))))))))..)).))..)))))).(((.(....).))).... ( -32.06) >DroEre_CAF1 16295 115 + 1 GC---CAACUUCGCUGCCGACGUCGCUGCUUAUAAAUCUUCGACAUGAAGCGCGCAACAAAAAGCCGCAAAAAA--AGGAAGGCAGAAACAGCAAGCGGCGUGCAUUGAAAAUUGCUCUA ((---....((((.(((..(((((((((((......(((((........(((.((........)))))......--..))))).......))).))))))))))).))))....)).... ( -30.91) >DroYak_CAF1 16563 115 + 1 GC---CAACUUCGCUGCCGACGUCGCUGCUUAUAAAUCUUCGACAUGAAGCGCGCAACAAAAAGCCGCAAAAAA--AAGAAGGCAGAAACAGCAAGCGGCGUGCAUUGAAAAUUGCUCUA ((---....((((.(((..(((((((((((......(((((........(((.((........)))))......--..))))).......))).))))))))))).))))....)).... ( -32.01) >consensus GC___CAACUUCGCUGCCGGCGUCGCUGCUUAUAAAUCUUCGACAUGAAGC__GCAACAAAAAGCCGCAAAAAA__AAGAAGGCAGAAACAGCAAGCGGCGCGCAUUGAAAAUUGCUCUA .........((((.(((..(((((((((((.......((((.....)))).............(((...............)))......))).))))))))))).)))).......... (-27.96 = -27.48 + -0.48)

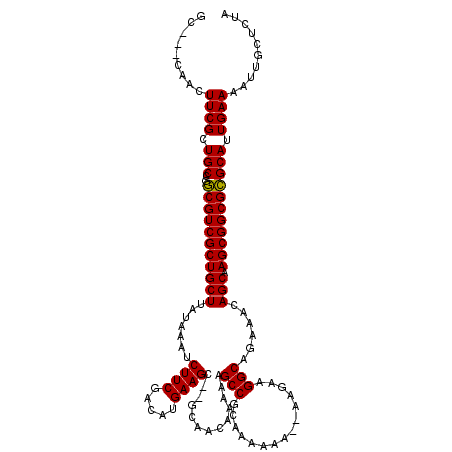
| Location | 12,077,679 – 12,077,799 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -45.00 |
| Consensus MFE | -44.40 |
| Energy contribution | -44.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12077679 120 + 27905053 AGGCAGAAACAGCAAGCGGCGCGCAUUGAAAAUUGCUCUAAUUCGCCAAAAACAGCUGAUUUGGUCGACGACGCCGCCGUCGCGGUGGCUUCCUGUCUCCGUCGCGACCGCUGCCUCCGC ((((((...((((....((((.(((.(....).))).......)))).......))))....(((((.(((((((((((...))))))).....(....)))))))))).)))))).... ( -45.00) >DroSec_CAF1 14432 120 + 1 AGGCAGAAACAGCAAGCGGCGCGCAUUGAAAAUUGCUCUAAUUCGCCAAAAACAGCUGAUUUGGUCGACGACGCCGCCGUCGCGGUGGCUUCCUGUCUCCGUCGCGACCGCUGCCUCCGC ((((((...((((....((((.(((.(....).))).......)))).......))))....(((((.(((((((((((...))))))).....(....)))))))))).)))))).... ( -45.00) >DroSim_CAF1 14626 120 + 1 AGGCAGAAACAGCAAGCGGCGCGCAUUGAAAAUUGCUCUAAUUCGCCAAAAACAGCUGAUUUGGUCGACGACGCCGCCGUCGCGGUGGCUUCCUGUCUCCGUCGCGACCGCUGCCUCCGC ((((((...((((....((((.(((.(....).))).......)))).......))))....(((((.(((((((((((...))))))).....(....)))))))))).)))))).... ( -45.00) >consensus AGGCAGAAACAGCAAGCGGCGCGCAUUGAAAAUUGCUCUAAUUCGCCAAAAACAGCUGAUUUGGUCGACGACGCCGCCGUCGCGGUGGCUUCCUGUCUCCGUCGCGACCGCUGCCUCCGC ((((((...((((....((((.(((........))).......)))).......))))....(((((.(((((((((((...))))))).....(....)))))))))).)))))).... (-44.40 = -44.40 + -0.00)



| Location | 12,077,679 – 12,077,799 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -54.80 |
| Consensus MFE | -54.80 |
| Energy contribution | -54.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12077679 120 - 27905053 GCGGAGGCAGCGGUCGCGACGGAGACAGGAAGCCACCGCGACGGCGGCGUCGUCGACCAAAUCAGCUGUUUUUGGCGAAUUAGAGCAAUUUUCAAUGCGCGCCGCUUGCUGUUUCUGCCU ((((((((((((((((((..(....).((...))..))))))((((((((..(((.(((((.........))))))))......(((........))))))))))))))))))))))).. ( -54.80) >DroSec_CAF1 14432 120 - 1 GCGGAGGCAGCGGUCGCGACGGAGACAGGAAGCCACCGCGACGGCGGCGUCGUCGACCAAAUCAGCUGUUUUUGGCGAAUUAGAGCAAUUUUCAAUGCGCGCCGCUUGCUGUUUCUGCCU ((((((((((((((((((..(....).((...))..))))))((((((((..(((.(((((.........))))))))......(((........))))))))))))))))))))))).. ( -54.80) >DroSim_CAF1 14626 120 - 1 GCGGAGGCAGCGGUCGCGACGGAGACAGGAAGCCACCGCGACGGCGGCGUCGUCGACCAAAUCAGCUGUUUUUGGCGAAUUAGAGCAAUUUUCAAUGCGCGCCGCUUGCUGUUUCUGCCU ((((((((((((((((((..(....).((...))..))))))((((((((..(((.(((((.........))))))))......(((........))))))))))))))))))))))).. ( -54.80) >consensus GCGGAGGCAGCGGUCGCGACGGAGACAGGAAGCCACCGCGACGGCGGCGUCGUCGACCAAAUCAGCUGUUUUUGGCGAAUUAGAGCAAUUUUCAAUGCGCGCCGCUUGCUGUUUCUGCCU ((((((((((((((((((..(....).((...))..))))))((((((((..(((.(((((.........))))))))......(((........))))))))))))))))))))))).. (-54.80 = -54.80 + 0.00)



| Location | 12,077,719 – 12,077,824 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -45.00 |
| Consensus MFE | -45.00 |
| Energy contribution | -45.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12077719 105 - 27905053 GUGGGGACGUCGAUUGGCAGCAGAGGCGGAGGCAGCGGUCGCGACGGAGACAGGAAGCCACCGCGACGGCGGCGUCGUCGACCAAAUCAGCUGUUUUUGGCGAAU .......((((((..((((((.(..((....))..)((((((((((....).....(((.(((...))).))))))).)))))......)))))).))))))... ( -45.00) >DroSec_CAF1 14472 105 - 1 GUGGGGACGUCGAUUGGCAGCAGAGGCGGAGGCAGCGGUCGCGACGGAGACAGGAAGCCACCGCGACGGCGGCGUCGUCGACCAAAUCAGCUGUUUUUGGCGAAU .......((((((..((((((.(..((....))..)((((((((((....).....(((.(((...))).))))))).)))))......)))))).))))))... ( -45.00) >DroSim_CAF1 14666 105 - 1 GUGGGGACGUCGAUUGGCAGCAGAGGCGGAGGCAGCGGUCGCGACGGAGACAGGAAGCCACCGCGACGGCGGCGUCGUCGACCAAAUCAGCUGUUUUUGGCGAAU .......((((((..((((((.(..((....))..)((((((((((....).....(((.(((...))).))))))).)))))......)))))).))))))... ( -45.00) >consensus GUGGGGACGUCGAUUGGCAGCAGAGGCGGAGGCAGCGGUCGCGACGGAGACAGGAAGCCACCGCGACGGCGGCGUCGUCGACCAAAUCAGCUGUUUUUGGCGAAU .......((((((..((((((.(..((....))..)((((((((((....).....(((.(((...))).))))))).)))))......)))))).))))))... (-45.00 = -45.00 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:32 2006