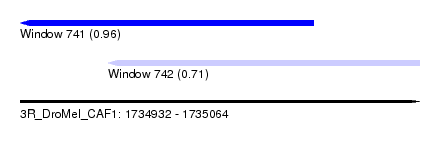
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,734,932 – 1,735,064 |
| Length | 132 |
| Max. P | 0.956770 |

| Location | 1,734,932 – 1,735,029 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 91.21 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -32.01 |
| Energy contribution | -32.65 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
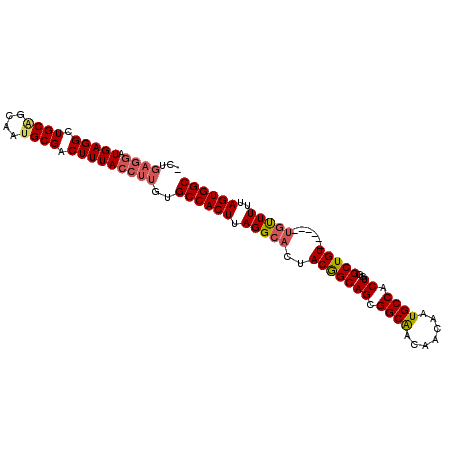
>3R_DroMel_CAF1 1734932 97 - 27905053 -CUGAGGAUGAGGCUGCAGCAAUGCGACUUUACCUUGUGCCACUUAGGAACUACGGCAGCGGCAACAACAAUGCCACUGGGCUGU------UGUUUUUAGUGGC -..((((.(((((.((((....)))).)))))))))..((((((.((((((.(((((((.((((.......)))).))..)))))------.)))))))))))) ( -39.80) >DroSec_CAF1 44072 97 - 1 -CUGAGGAUGAGGCUGCAGCAAUGCGACUUUACCUUGUGCCACUUAGGCACUACGGCAGCGGCAACAACAAUGCCACUGGGCUGU------UGUUUUUAGUGGC -..((((.(((((.((((....)))).)))))))))..((((((.(((((..(((((((.((((.......)))).))..)))))------)))))..)))))) ( -37.10) >DroSim_CAF1 45237 97 - 1 -CUGAGGAUGAGGCUGCAGCAAUGCGACUUUACCUUGUGCCACUUAGGCACUACGGCAGCGGCAACAACAAUGCCACUGGGCUGU------UGUUUUUAGUGGC -..((((.(((((.((((....)))).)))))))))..((((((.(((((..(((((((.((((.......)))).))..)))))------)))))..)))))) ( -37.10) >DroEre_CAF1 43034 97 - 1 -GUGAGGAUGAGGCUGCAGCAAUGCGACUUUACCUUGUGCCACUUAGGCACUACGGCAGCGGCGGCAACAAUGCCACUGGGCUGU------UGUUUUUAGUGGC -(..(((.(((((.((((....)))).))))))))..)((((((.(((((..(((((..(((.((((....)))).))).)))))------)))))..)))))) ( -42.30) >DroYak_CAF1 44139 98 - 1 GAUGAGGAUGAGGCUGCAGCAAAGCGACUUUACCUUGUGCCACUUAGGCUCUACGGCAGGGGCAACAACAAUGCCACUGGGCUGU------UGUUUUUAGUGGC .(..(((.(((((.(((......))).))))))))..)((((((.((((...(((((.((((((.......)))).))..)))))------.))))..)))))) ( -36.40) >DroAna_CAF1 39486 97 - 1 -------CUGAGGCUGCGGCAAUGCGACUUUACCUUGUGCCACUUAGGCACUACAGCAGUGGCAACAACAAUGCCACUGUGCUGUUCUUGUUGCUUUUAGUGGC -------.(((((.((((....)))).)))))......((((((.((((((.((((((((((((.......))))))..))))))....).)))))..)))))) ( -36.00) >consensus _CUGAGGAUGAGGCUGCAGCAAUGCGACUUUACCUUGUGCCACUUAGGCACUACGGCAGCGGCAACAACAAUGCCACUGGGCUGU______UGUUUUUAGUGGC ...((((.(((((.((((....)))).)))))))))..((((((.(((((..(((((((.((((.......)))).))..)))))......)))))..)))))) (-32.01 = -32.65 + 0.64)

| Location | 1,734,961 – 1,735,064 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.32 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -25.41 |
| Energy contribution | -26.25 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
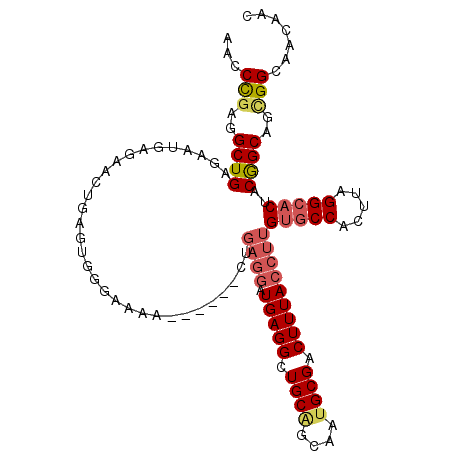
>3R_DroMel_CAF1 1734961 103 - 27905053 AACCCGAGGCUGAGAAUGAGAACUGAGUGGAAAAA------CUGAGGAUGAGGCUGCAGCAAUGCGACUUUACCUUGUGCCACUUAGGAACUACGGCAGCGGCAACAAC ...(((..((((((........((((((((....(------(.((((.(((((.((((....)))).))))))))))).))))))))...)).))))..)))....... ( -35.70) >DroSec_CAF1 44101 103 - 1 AACCCGAGGCUGAGAAUGAGAACUAAGUGGGAAAA------CUGAGGAUGAGGCUGCAGCAAUGCGACUUUACCUUGUGCCACUUAGGCACUACGGCAGCGGCAACAAC ...(((..((((((..((....((((((((....(------(.((((.(((((.((((....)))).))))))))))).)))))))).)))).))))..)))....... ( -36.20) >DroSim_CAF1 45266 103 - 1 AACCCGAGGCUGAGAAUGAGAACUAAGUGGGAAAA------CUGAGGAUGAGGCUGCAGCAAUGCGACUUUACCUUGUGCCACUUAGGCACUACGGCAGCGGCAACAAC ...(((..((((((..((....((((((((....(------(.((((.(((((.((((....)))).))))))))))).)))))))).)))).))))..)))....... ( -36.20) >DroEre_CAF1 43063 103 - 1 AACCCGAGGCUGAGAAUGAAAACUGAGUGGGAAAA------GUGAGGAUGAGGCUGCAGCAAUGCGACUUUACCUUGUGCCACUUAGGCACUACGGCAGCGGCGGCAAC ..((((..((((((..((....((((((((.....------(..(((.(((((.((((....)))).))))))))..).)))))))).)))).))))..))).)..... ( -37.60) >DroYak_CAF1 44168 109 - 1 AACCCGAGGCUGAGAAUGAAAACUGAGUGGGAAAACAGAGGAUGAGGAUGAGGCUGCAGCAAAGCGACUUUACCUUGUGCCACUUAGGCUCUACGGCAGGGGCAACAAC ..(((...(((((((..(....((((((((.(.....((((.(((((.((..((....))....)).))))))))).).)))))))).)))).)))).)))........ ( -34.50) >DroAna_CAF1 39521 78 - 1 CACCUGAGGCUGAAAU-------------------------------CUGAGGCUGCGGCAAUGCGACUUUACCUUGUGCCACUUAGGCACUACAGCAGUGGCAACAAC ........((((....-------------------------------.(((((.((((....)))).)))))....(((((.....)))))..))))..((....)).. ( -21.80) >consensus AACCCGAGGCUGAGAAUGAGAACUGAGUGGGAAAA______CUGAGGAUGAGGCUGCAGCAAUGCGACUUUACCUUGUGCCACUUAGGCACUACGGCAGCGGCAACAAC ...(((..((((...............................((((.(((((.((((....)))).)))))))))(((((.....)))))..))))..)))....... (-25.41 = -26.25 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:40 2006