
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,075,982 – 12,076,599 |
| Length | 617 |
| Max. P | 0.999964 |

| Location | 12,075,982 – 12,076,102 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -27.96 |
| Energy contribution | -28.60 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12075982 120 + 27905053 CUCCCUUUACCCCUCUGCGUUGCAGCGGCCCAUACUUUAUUUUUAACUGUGCGACUGUACUUCGUGUGUCGGUCGUUCGGCUCGUUCGCUUGUACAUGUUUUUACAUAUAUACGAACGGA ...........((.((((...)))).))..................(((.((((((((((.....))).))))))).))).(((((((..((((.((((....)))).))))))))))). ( -32.00) >DroSec_CAF1 12755 118 + 1 CUCCCUUUACCCCUCAGCGUUGCACCGGCCCACACUUUAUUUUUAACUGUGCGACUGUACUUCGU--GUCGGUCGUUCGGCUCGUUCGCUUGGACAUGUUUUUACAUAUAUACGAACGGA .(((.(((...((..((((....((.((((((((.((.......)).))))((((((.((.....--))))))))...)))).)).)))).))..((((....))))......))).))) ( -26.30) >DroSim_CAF1 12974 120 + 1 CUCCCUUUACCCCUCCGCGUUGCAGCGGCCCACACUUUAUUUUUAACUGUGCGACUGUACUUCGUGUGUCGGUCGUUCGGCUCGUUCGCUUGUACAUGUUUUUACAUAUAUACGAACGGA .............((((.((((.((((((((((((.(((....)))..((((....))))...)))))..))))))))))).)(((((..((((.((((....)))).)))))))))))) ( -32.70) >DroEre_CAF1 14619 119 + 1 CUCCCUUU-CCCCUUCGCGUUGCAGCGGCCCACACUUUAUUUUUAACUGUGCGACUGUACUUCGUGUGUCGGUCGUUCGGCUCGUUCGCUUGUACAUGUUUAUACAUAUAUACGAACGGA .......(-((.....(.((((.((((((((((((.(((....)))..((((....))))...)))))..))))))))))).)(((((..((((.((((....)))).)))))))))))) ( -31.90) >DroYak_CAF1 14890 119 + 1 CUCCCUUU-CCCCUUCGCGUUGCAGCGGCCCAUACUUUAUUUUUAACUGUGCGACUGUACUUCGUGUGUCGGUCGUUCGGCUCGUUCGCUUGUACAUGUUUGUACAUAUAUACGAACGGA ........-.....((((......))))..................(((.((((((((((.....))).))))))).))).(((((((..((((((....))))))......))))))). ( -32.20) >consensus CUCCCUUUACCCCUCCGCGUUGCAGCGGCCCACACUUUAUUUUUAACUGUGCGACUGUACUUCGUGUGUCGGUCGUUCGGCUCGUUCGCUUGUACAUGUUUUUACAUAUAUACGAACGGA ..................((((.((((((((((((.(((....)))..((((....))))...)))))..)))))))))))(((((((..((((.((((....)))).))))))))))). (-27.96 = -28.60 + 0.64)

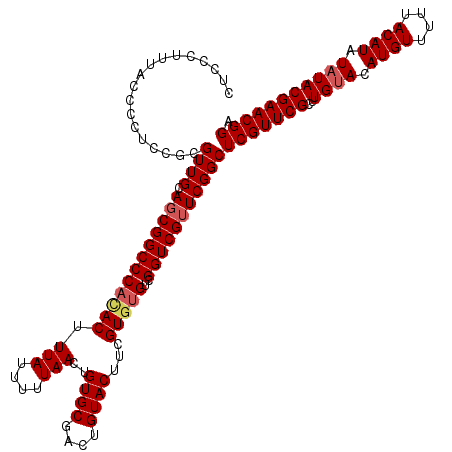
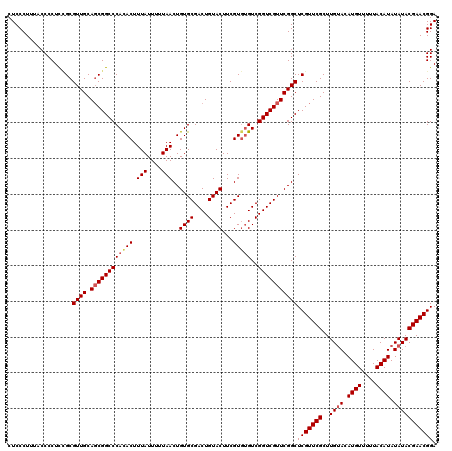
| Location | 12,076,022 – 12,076,142 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.75 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -34.48 |
| Energy contribution | -34.72 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076022 120 + 27905053 UUUUAACUGUGCGACUGUACUUCGUGUGUCGGUCGUUCGGCUCGUUCGCUUGUACAUGUUUUUACAUAUAUACGAACGGAGCGUCUGUAUGUAUGCGGUGAGUCGAGUCAGCAACAUUGG ......(((..(((((.((((.((((..((((.(((((....((((((..((((.((((....)))).))))))))))))))).))).)..)))).)))))))))...)))......... ( -38.00) >DroSec_CAF1 12795 118 + 1 UUUUAACUGUGCGACUGUACUUCGU--GUCGGUCGUUCGGCUCGUUCGCUUGGACAUGUUUUUACAUAUAUACGAACGGAGCGUCUGUAUGUAUGCGGUGAGUCGAGUCAGCAACAUUGG .......((((((((((.((.....--)))))))(((((((((((((....))))......(..((((((((((.(((...))).))))))))))..).)))))))).).)).))).... ( -36.30) >DroSim_CAF1 13014 120 + 1 UUUUAACUGUGCGACUGUACUUCGUGUGUCGGUCGUUCGGCUCGUUCGCUUGUACAUGUUUUUACAUAUAUACGAACGGAGCGUCUGUAUGUAUGCGGUGAGUCGAGUCAGCAACAUUGG ......(((..(((((.((((.((((..((((.(((((....((((((..((((.((((....)))).))))))))))))))).))).)..)))).)))))))))...)))......... ( -38.00) >DroEre_CAF1 14658 120 + 1 UUUUAACUGUGCGACUGUACUUCGUGUGUCGGUCGUUCGGCUCGUUCGCUUGUACAUGUUUAUACAUAUAUACGAACGGAGCGUGUGUAUGUAUGCGGUGAGUCGAGUCAGCAACAUUGG .......(((((((((((((.....))).)))))))..((((((((((((.((........(((((((((((((.......))))))))))))))))))))).)))))).)))....... ( -40.90) >DroYak_CAF1 14929 120 + 1 UUUUAACUGUGCGACUGUACUUCGUGUGUCGGUCGUUCGGCUCGUUCGCUUGUACAUGUUUGUACAUAUAUACGAACGGAGUGUCUGUAUGUAUGCGGUGAGUCGAGUCAGCAACAUUGG .......(((((((((((((.....))).)))))))..((((((((((((((((((....))))))..((((((.((((.....)))).)))))).)))))).)))))).)))....... ( -41.40) >consensus UUUUAACUGUGCGACUGUACUUCGUGUGUCGGUCGUUCGGCUCGUUCGCUUGUACAUGUUUUUACAUAUAUACGAACGGAGCGUCUGUAUGUAUGCGGUGAGUCGAGUCAGCAACAUUGG .......(((((((((((((.....))).)))))))..((((((((((((..............((((((((((.(((...))).)))))))))).)))))).)))))).)))....... (-34.48 = -34.72 + 0.24)

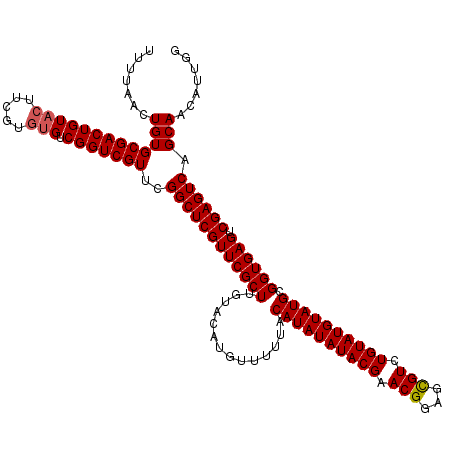

| Location | 12,076,022 – 12,076,142 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.75 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -26.90 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076022 120 - 27905053 CCAAUGUUGCUGACUCGACUCACCGCAUACAUACAGACGCUCCGUUCGUAUAUAUGUAAAAACAUGUACAAGCGAACGAGCCGAACGACCGACACACGAAGUACAGUCGCACAGUUAAAA ....(((.(((((((((.....................((((.((((((.(((((((....)))))))...))))))))))((......)).....)).))).)))).)))......... ( -28.60) >DroSec_CAF1 12795 118 - 1 CCAAUGUUGCUGACUCGACUCACCGCAUACAUACAGACGCUCCGUUCGUAUAUAUGUAAAAACAUGUCCAAGCGAACGAGCCGAACGACCGAC--ACGAAGUACAGUCGCACAGUUAAAA ....(((.(((((((((.....................((((.((((((..((((((....))))))....))))))))))((......))..--.)).))).)))).)))......... ( -27.00) >DroSim_CAF1 13014 120 - 1 CCAAUGUUGCUGACUCGACUCACCGCAUACAUACAGACGCUCCGUUCGUAUAUAUGUAAAAACAUGUACAAGCGAACGAGCCGAACGACCGACACACGAAGUACAGUCGCACAGUUAAAA ....(((.(((((((((.....................((((.((((((.(((((((....)))))))...))))))))))((......)).....)).))).)))).)))......... ( -28.60) >DroEre_CAF1 14658 120 - 1 CCAAUGUUGCUGACUCGACUCACCGCAUACAUACACACGCUCCGUUCGUAUAUAUGUAUAAACAUGUACAAGCGAACGAGCCGAACGACCGACACACGAAGUACAGUCGCACAGUUAAAA ....(((.(((((((((.....................((((.((((((.(((((((....)))))))...))))))))))((......)).....)).))).)))).)))......... ( -28.60) >DroYak_CAF1 14929 120 - 1 CCAAUGUUGCUGACUCGACUCACCGCAUACAUACAGACACUCCGUUCGUAUAUAUGUACAAACAUGUACAAGCGAACGAGCCGAACGACCGACACACGAAGUACAGUCGCACAGUUAAAA ........((((..(((.(((..(((....((((..((.....))..))))...((((((....)))))).)))...))).))).((((.(((.......)).).))))..))))..... ( -26.30) >consensus CCAAUGUUGCUGACUCGACUCACCGCAUACAUACAGACGCUCCGUUCGUAUAUAUGUAAAAACAUGUACAAGCGAACGAGCCGAACGACCGACACACGAAGUACAGUCGCACAGUUAAAA ....(((.(((((((((.....................((((.((((((.(((((((....)))))))...))))))))))((......)).....)).))).)))).)))......... (-26.90 = -27.30 + 0.40)

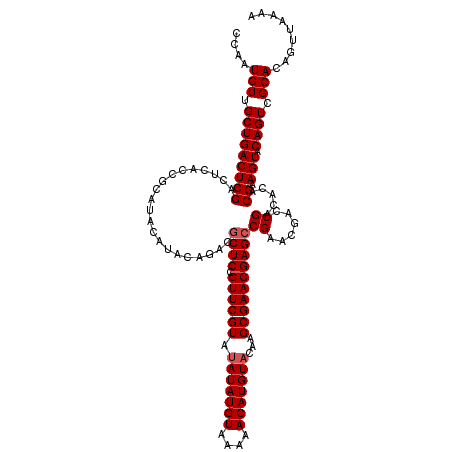
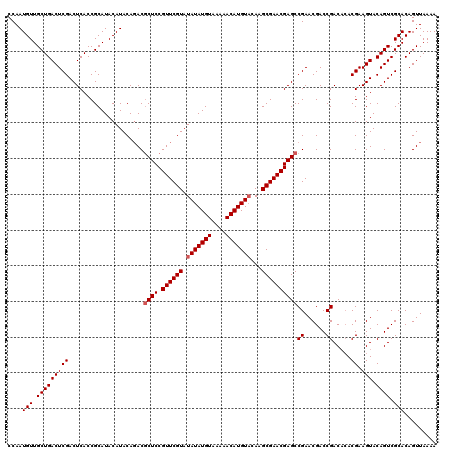
| Location | 12,076,062 – 12,076,182 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -22.96 |
| Energy contribution | -23.16 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076062 120 - 27905053 CCAUGCCUACAUAUGUAUGUAUGUACAUCCACAGCUACAUCCAAUGUUGCUGACUCGACUCACCGCAUACAUACAGACGCUCCGUUCGUAUAUAUGUAAAAACAUGUACAAGCGAACGAG .............(((((((((((.......((((.((((...)))).))))....(......)))))))))))).......(((((((.(((((((....)))))))...))))))).. ( -34.00) >DroSec_CAF1 12833 112 - 1 CCAUGCCUACAUAU--------GUACAUCCACAGCUACAUCCAAUGUUGCUGACUCGACUCACCGCAUACAUACAGACGCUCCGUUCGUAUAUAUGUAAAAACAUGUCCAAGCGAACGAG ....((.....(((--------(((......((((.((((...)))).))))...((......))..)))))).....))..(((((((..((((((....))))))....))))))).. ( -23.90) >DroSim_CAF1 13054 112 - 1 CCAUGCCUACAUAU--------GUACAUCCACAGCUACAUCCAAUGUUGCUGACUCGACUCACCGCAUACAUACAGACGCUCCGUUCGUAUAUAUGUAAAAACAUGUACAAGCGAACGAG ....((.....(((--------(((......((((.((((...)))).))))...((......))..)))))).....))..(((((((.(((((((....)))))))...))))))).. ( -25.50) >DroEre_CAF1 14698 112 - 1 CCAUGCAUACAUAC--------GUACAUCCACAGCUACAUCCAAUGUUGCUGACUCGACUCACCGCAUACAUACACACGCUCCGUUCGUAUAUAUGUAUAAACAUGUACAAGCGAACGAG ..((((.(((....--------)))......((((.((((...)))).))))............))))..............(((((((.(((((((....)))))))...))))))).. ( -25.60) >DroYak_CAF1 14969 112 - 1 CCAUGCAUACAUAU--------GUACAUCCACAGCUACAUCCAAUGUUGCUGACUCGACUCACCGCAUACAUACAGACACUCCGUUCGUAUAUAUGUACAAACAUGUACAAGCGAACGAG ..((((.(((....--------)))......((((.((((...)))).))))............))))..............(((((((.(((((((....)))))))...))))))).. ( -25.90) >consensus CCAUGCCUACAUAU________GUACAUCCACAGCUACAUCCAAUGUUGCUGACUCGACUCACCGCAUACAUACAGACGCUCCGUUCGUAUAUAUGUAAAAACAUGUACAAGCGAACGAG ..((((.........................((((.((((...)))).))))....(......)))))..............(((((((.(((((((....)))))))...))))))).. (-22.96 = -23.16 + 0.20)


| Location | 12,076,142 – 12,076,254 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.54 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076142 112 + 27905053 AUGUAGCUGUGGAUGUACAUACAUACAUAUGUAGGCAUGGGAUGUGGAGUGGAGUCUUUGUUGGUUGGGGCUGUAUGUACUUA--------CAUACAUAUAUACCCACAGUUUUUUUCAU ....((((((((.((((..(((((...((((....))))..)))))......((((((........))))))((((((....)--------)))))...)))).))))))))........ ( -28.90) >DroSec_CAF1 12913 104 + 1 AUGUAGCUGUGGAUGUAC--------AUAUGUAGGCAUGGGAUGUGGAGUGGAGUCUUUGUUGGUUGGGGCUGUAUGUACAUU--------CAAACAUAUAUACCCACAGUUUUUUCCAU ....((((((((..(((.--------((((((.......(((((((..(((.((((((........)))))).))).))))))--------)..)))))).)))))))))))........ ( -30.60) >DroSim_CAF1 13134 104 + 1 AUGUAGCUGUGGAUGUAC--------AUAUGUAGGCAUGGGAUGUGGAGUGGAGUCUUUGUUGGUUGGGGCUGUAUGUACAUU--------CAAACAUAUAUACCCACAGUUUUUUCCAU ....((((((((..(((.--------((((((.......(((((((..(((.((((((........)))))).))).))))))--------)..)))))).)))))))))))........ ( -30.60) >DroEre_CAF1 14778 112 + 1 AUGUAGCUGUGGAUGUAC--------GUAUGUAUGCAUGGGAUGUGGAGUGGAGUCUUUGUUGGUUGGGGCUGUAUGUACAAACAUACAUUCGUACAUAUAUACCCACAGUUUUUUCCAU ....((((((((..(((.--------.((((((((.(((..((((...(((.((((((........)))))).)))......)))).))).))))))))..)))))))))))........ ( -36.00) >DroYak_CAF1 15049 108 + 1 AUGUAGCUGUGGAUGUAC--------AUAUGUAUGCAUGGGAUGUGGAGUGGAGUCUUUGUUGGUUGGGGCUAUAUGUACAAACAUA----CGUACAUAUAUACCCACAGUUUUUUCCAU ....((((((((..(((.--------(((((((((.(((...((((.(....((((((........))))))...).))))..))).----))))))))).)))))))))))........ ( -33.50) >consensus AUGUAGCUGUGGAUGUAC________AUAUGUAGGCAUGGGAUGUGGAGUGGAGUCUUUGUUGGUUGGGGCUGUAUGUACAUA________CAUACAUAUAUACCCACAGUUUUUUCCAU ....((((((((.((((..........((((....))))...((((..(((.((((((........)))))).))).))))..................)))).))))))))........ (-23.34 = -23.54 + 0.20)



| Location | 12,076,182 – 12,076,292 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.55 |
| Mean single sequence MFE | -17.48 |
| Consensus MFE | -13.04 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076182 110 - 27905053 CGCUUGGUAAACACAAAAUUACAAAAACAAA--AAAUACCAUGAAAAAAACUGUGGGUAUAUAUGUAUG--------UAAGUACAUACAGCCCCAACCAACAAAGACUCCACUCCACAUC .(.(((((.......................--........(....)....((.((((...(((((((.--------...)))))))..))))))))))))................... ( -14.70) >DroSec_CAF1 12945 110 - 1 CGCUUGGUAAACACAAAAUUACAAAAACAAA--AAAUACCAUGGAAAAAACUGUGGGUAUAUAUGUUUG--------AAUGUACAUACAGCCCCAACCAACAAAGACUCCACUCCACAUC .(.(((((.......................--.....((((((......))))))((((((.......--------.))))))...........))))))................... ( -14.50) >DroSim_CAF1 13166 110 - 1 CGCUUGGUAAACACAAAAUUACAAAAACAAA--AAAUACCAUGGAAAAAACUGUGGGUAUAUAUGUUUG--------AAUGUACAUACAGCCCCAACCAACAAAGACUCCACUCCACAUC .(.(((((.......................--.....((((((......))))))((((((.......--------.))))))...........))))))................... ( -14.50) >DroEre_CAF1 14810 119 - 1 CGCUUGGUAAACACAAAAUUACAAAAACAAA-AAAAUACCAUGGAAAAAACUGUGGGUAUAUAUGUACGAAUGUAUGUUUGUACAUACAGCCCCAACCAACAAAGACUCCACUCCACAUC .(.(((((.......................-......((((((......)))))).....(((((((((((....)))))))))))........))))))................... ( -21.50) >DroYak_CAF1 15081 116 - 1 CGCUUGGUAAACACAAAAUUACAAAAACAAAAAAAAUACCAUGGAAAAAACUGUGGGUAUAUAUGUACG----UAUGUUUGUACAUAUAGCCCCAACCAACAAAGACUCCACUCCACAUC ....(((((...........................)))))((((........((((..((((((((((----......))))))))))..))))............))))......... ( -22.18) >consensus CGCUUGGUAAACACAAAAUUACAAAAACAAA__AAAUACCAUGGAAAAAACUGUGGGUAUAUAUGUAUG________UAUGUACAUACAGCCCCAACCAACAAAGACUCCACUCCACAUC .(.(((((..............................((((((......)))))).....(((((((............)))))))........))))))................... (-13.04 = -13.40 + 0.36)


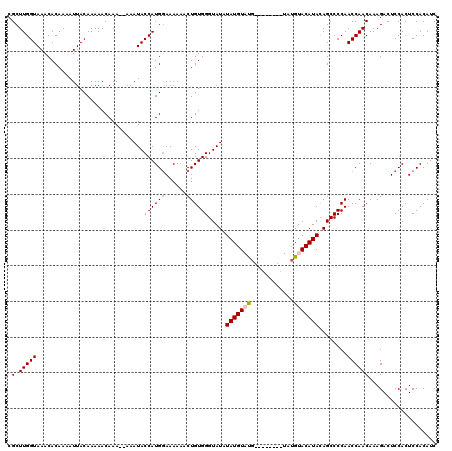
| Location | 12,076,222 – 12,076,332 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.21 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.54 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076222 110 + 27905053 UUA--------CAUACAUAUAUACCCACAGUUUUUUUCAUGGUAUUU--UUUGUUUUUGUAAUUUUGUGUUUACCAAGCGGCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUGU ...--------...............((((((....(((.(((....--...................((((((((((...))))))))))((((....))))))).))).))))))... ( -24.90) >DroSec_CAF1 12985 110 + 1 AUU--------CAAACAUAUAUACCCACAGUUUUUUCCAUGGUAUUU--UUUGUUUUUGUAAUUUUGUGUUUACCAAGCGGCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUUU ...--------...............((((((...((...(((....--...................((((((((((...))))))))))((((....)))))))..)).))))))... ( -22.90) >DroSim_CAF1 13206 110 + 1 AUU--------CAAACAUAUAUACCCACAGUUUUUUCCAUGGUAUUU--UUUGUUUUUGUAAUUUUGUGUUUACCAAGCGGCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUGU ...--------...............((((((...((...(((....--...................((((((((((...))))))))))((((....)))))))..)).))))))... ( -22.90) >DroEre_CAF1 14850 119 + 1 AAACAUACAUUCGUACAUAUAUACCCACAGUUUUUUCCAUGGUAUUUU-UUUGUUUUUGUAAUUUUGUGUUUACCAAGCGGCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUGU ..........................((((((...((...(((.....-...................((((((((((...))))))))))((((....)))))))..)).))))))... ( -22.90) >DroYak_CAF1 15121 116 + 1 AAACAUA----CGUACAUAUAUACCCACAGUUUUUUCCAUGGUAUUUUUUUUGUUUUUGUAAUUUUGUGUUUACCAAGCGGCUUGGUGAAUGUACGUACGUACGCCUCGAUAGCUGUUGU .((((((----(((((....(((((...............)))))....................((..(((((((((...)))))))))..)).))))))).((.......)))))).. ( -23.56) >consensus AUA________CAUACAUAUAUACCCACAGUUUUUUCCAUGGUAUUU__UUUGUUUUUGUAAUUUUGUGUUUACCAAGCGGCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUGU ..........................((((((...((...(((.........................((((((((((...))))))))))((((....)))))))..)).))))))... (-22.70 = -22.54 + -0.16)


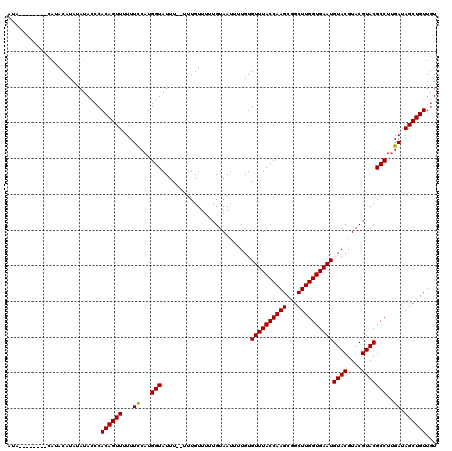
| Location | 12,076,222 – 12,076,332 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.21 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -22.64 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.95 |
| SVM RNA-class probability | 0.999964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076222 110 - 27905053 ACAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGCCGCUUGGUAAACACAAAAUUACAAAAACAAA--AAAUACCAUGAAAAAAACUGUGGGUAUAUAUGUAUG--------UAA ...............(((((((((..(((....((((((...)))))).......................--.....(((((........))))))))..))))))))--------).. ( -22.60) >DroSec_CAF1 12985 110 - 1 AAAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGCCGCUUGGUAAACACAAAAUUACAAAAACAAA--AAAUACCAUGGAAAAAACUGUGGGUAUAUAUGUUUG--------AAU ......(((.....)))((((....))))((((((((((...))))))................(((((..--.....((((((......)))))).......))))))--------))) ( -21.54) >DroSim_CAF1 13206 110 - 1 ACAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGCCGCUUGGUAAACACAAAAUUACAAAAACAAA--AAAUACCAUGGAAAAAACUGUGGGUAUAUAUGUUUG--------AAU ......(((.....)))((((....))))((((((((((...))))))................(((((..--.....((((((......)))))).......))))))--------))) ( -21.54) >DroEre_CAF1 14850 119 - 1 ACAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGCCGCUUGGUAAACACAAAAUUACAAAAACAAA-AAAAUACCAUGGAAAAAACUGUGGGUAUAUAUGUACGAAUGUAUGUUU .............((((((((((.(((((((..((((((...)))))).......................-......((((((......))))))......))))))).)))))))))) ( -29.40) >DroYak_CAF1 15121 116 - 1 ACAACAGCUAUCGAGGCGUACGUACGUACAUUCACCAAGCCGCUUGGUAAACACAAAAUUACAAAAACAAAAAAAAUACCAUGGAAAAAACUGUGGGUAUAUAUGUACG----UAUGUUU .............(((((((((((((((.....((((((...))))))..............................((((((......)))))).....))))))))----))))))) ( -29.20) >consensus ACAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGCCGCUUGGUAAACACAAAAUUACAAAAACAAA__AAAUACCAUGGAAAAAACUGUGGGUAUAUAUGUAUG________UAU ......(((.....)))(((((((..(((....((((((...))))))..............................((((((......)))))))))..)))))))............ (-22.64 = -23.00 + 0.36)


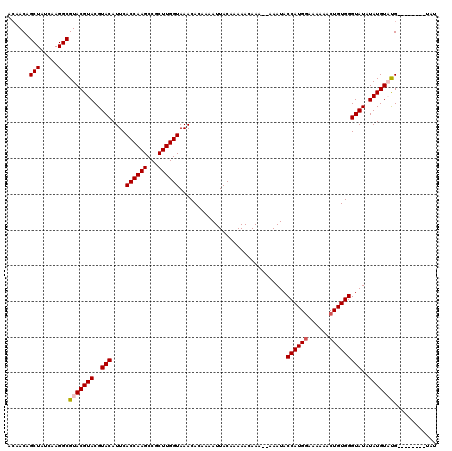
| Location | 12,076,254 – 12,076,372 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.49 |
| Mean single sequence MFE | -40.76 |
| Consensus MFE | -40.58 |
| Energy contribution | -40.58 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.11 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.58 |
| SVM RNA-class probability | 0.999923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076254 118 + 27905053 GGUAUUU--UUUGUUUUUGUAAUUUUGUGUUUACCAAGCGGCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUGUUGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGC ((((...--.........((((........))))..(((((((.((((.((((....)))).)))).....)))))))..))))..(((((((((((((........))))))))))))) ( -40.70) >DroSec_CAF1 13017 118 + 1 GGUAUUU--UUUGUUUUUGUAAUUUUGUGUUUACCAAGCGGCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUUUUGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGC ((((...--.........((((........)))).((((((((.((((.((((....)))).)))).....)))))))).))))..(((((((((((((........))))))))))))) ( -41.00) >DroSim_CAF1 13238 118 + 1 GGUAUUU--UUUGUUUUUGUAAUUUUGUGUUUACCAAGCGGCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUGUUGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGC ((((...--.........((((........))))..(((((((.((((.((((....)))).)))).....)))))))..))))..(((((((((((((........))))))))))))) ( -40.70) >DroEre_CAF1 14890 119 + 1 GGUAUUUU-UUUGUUUUUGUAAUUUUGUGUUUACCAAGCGGCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUGUUGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGC ((((....-.........((((........))))..(((((((.((((.((((....)))).)))).....)))))))..))))..(((((((((((((........))))))))))))) ( -40.70) >DroYak_CAF1 15157 120 + 1 GGUAUUUUUUUUGUUUUUGUAAUUUUGUGUUUACCAAGCGGCUUGGUGAAUGUACGUACGUACGCCUCGAUAGCUGUUGUUGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGC ((((..............((((........))))..(((((((.((((.((((....)))).)))).....)))))))..))))..(((((((((((((........))))))))))))) ( -40.70) >consensus GGUAUUU__UUUGUUUUUGUAAUUUUGUGUUUACCAAGCGGCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUGUUGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGC ((((..............((((........))))..(((((((.((((.((((....)))).)))).....)))))))..))))..(((((((((((((........))))))))))))) (-40.58 = -40.58 + -0.00)

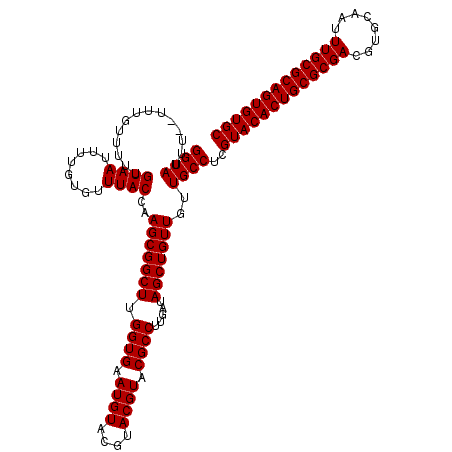
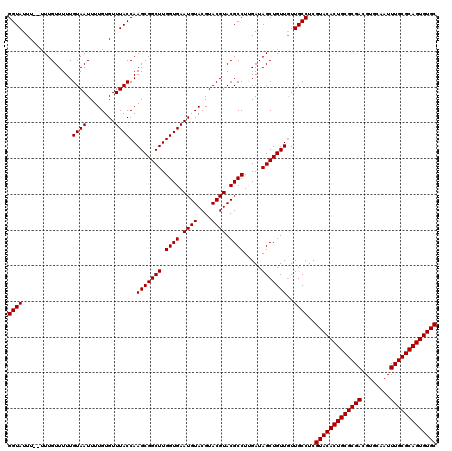
| Location | 12,076,254 – 12,076,372 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.49 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -30.30 |
| Energy contribution | -30.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076254 118 - 27905053 GCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCAACAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGCCGCUUGGUAAACACAAAAUUACAAAAACAAA--AAAUACC (((((((((((............)))))))))....(((.......(((.....)))((((....)))).........)))))...((((........)))).........--....... ( -30.30) >DroSec_CAF1 13017 118 - 1 GCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCAAAAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGCCGCUUGGUAAACACAAAAUUACAAAAACAAA--AAAUACC (((((((((((............)))))))))....(((.......(((.....)))((((....)))).........)))))...((((........)))).........--....... ( -30.30) >DroSim_CAF1 13238 118 - 1 GCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCAACAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGCCGCUUGGUAAACACAAAAUUACAAAAACAAA--AAAUACC (((((((((((............)))))))))....(((.......(((.....)))((((....)))).........)))))...((((........)))).........--....... ( -30.30) >DroEre_CAF1 14890 119 - 1 GCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCAACAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGCCGCUUGGUAAACACAAAAUUACAAAAACAAA-AAAAUACC (((((((((((............)))))))))....(((.......(((.....)))((((....)))).........)))))...((((........)))).........-........ ( -30.30) >DroYak_CAF1 15157 120 - 1 GCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCAACAACAGCUAUCGAGGCGUACGUACGUACAUUCACCAAGCCGCUUGGUAAACACAAAAUUACAAAAACAAAAAAAAUACC (((((((((((............))))))))).((((((.......))).)))..))((((....))))....((((((...))))))................................ ( -31.20) >consensus GCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCAACAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGCCGCUUGGUAAACACAAAAUUACAAAAACAAA__AAAUACC (((((((((((............)))))))))....(((.......(((.....)))((((....)))).........)))))...((((........)))).................. (-30.30 = -30.30 + 0.00)

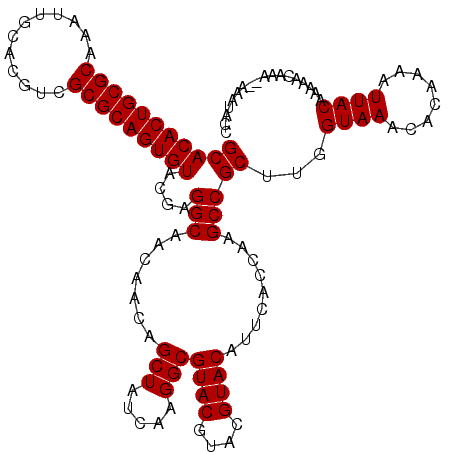

| Location | 12,076,292 – 12,076,412 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -46.72 |
| Consensus MFE | -46.36 |
| Energy contribution | -46.40 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.18 |
| SVM RNA-class probability | 0.999827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076292 120 + 27905053 GCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUGUUGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGCUGUCGCUCUGACAAUUGUUAACGGCAAUUGUUGUUGCUCG ((..((((.((((....)))).))))..((((((....((((((..(((((((((((((........)))))))))))))((((.....)))).........)))))).))))))))... ( -45.80) >DroSec_CAF1 13055 120 + 1 GCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUUUUGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGCUGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCG ....((((.((((....)))).))))..((((((.......))...(((((((((((((........)))))))))))))))))((..((((((((((.....))))))))))..))... ( -46.50) >DroSim_CAF1 13276 120 + 1 GCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUGUUGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGCUGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCG ((..((((.((((....)))).))))..((((((....((((((..(((((((((((((........)))))))))))))(((((...))))).........)))))).))))))))... ( -47.00) >DroEre_CAF1 14929 120 + 1 GCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUGUUGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGCUGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCG ((..((((.((((....)))).))))..((((((....((((((..(((((((((((((........)))))))))))))(((((...))))).........)))))).))))))))... ( -47.00) >DroYak_CAF1 15197 120 + 1 GCUUGGUGAAUGUACGUACGUACGCCUCGAUAGCUGUUGUUGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGCUGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCG ....((((.((((....)))).)))).(((((((....((((((..(((((((((((((........)))))))))))))(((((...))))).........)))))).))))))).... ( -47.30) >consensus GCUUGGUGAAUGUACGUACGUACGCCUUGAUAGCUGUUGUUGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGCUGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCG ((..((((.((((....)))).))))..((((((....((((((..(((((((((((((........)))))))))))))(((((...))))).........)))))).))))))))... (-46.36 = -46.40 + 0.04)

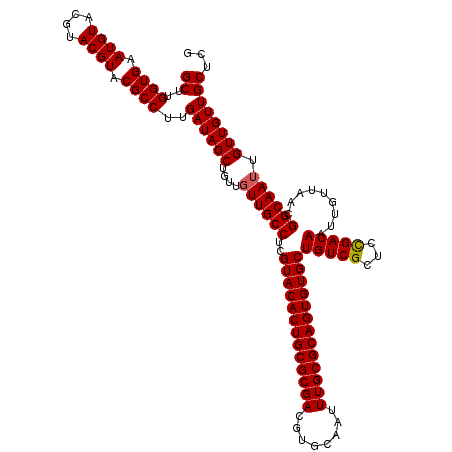
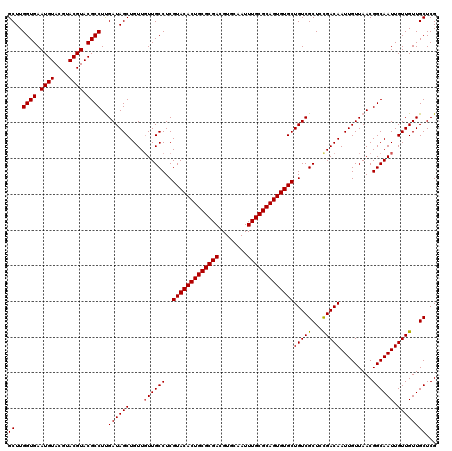
| Location | 12,076,292 – 12,076,412 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -34.74 |
| Consensus MFE | -33.36 |
| Energy contribution | -33.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076292 120 - 27905053 CGAGCAACAACAAUUGCCGUUAACAAUUGUCAGAGCGACAGCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCAACAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGC .((((....(((((((.......)))))))......((.((((((((((((............)))))))))....(....)....))).))...))((((....))))..))....... ( -33.10) >DroSec_CAF1 13055 120 - 1 CGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACAGCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCAAAAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGC ...((........(((((........((((((...)))))).(((((((((............)))))))))....))))).....(((.....)))((((....)))).........)) ( -34.00) >DroSim_CAF1 13276 120 - 1 CGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACAGCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCAACAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGC ...((..(.(((((((.......))))))).)..))((.((((((((((((............)))))))))....(....)....))).)).....((((....))))........... ( -35.00) >DroEre_CAF1 14929 120 - 1 CGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACAGCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCAACAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGC ...((..(.(((((((.......))))))).)..))((.((((((((((((............)))))))))....(....)....))).)).....((((....))))........... ( -35.00) >DroYak_CAF1 15197 120 - 1 CGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACAGCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCAACAACAGCUAUCGAGGCGUACGUACGUACAUUCACCAAGC ...((..(.(((((((.......))))))).)..))((.((((((((((((............)))))))))....(....)....))).))(((..((((....)))).)))....... ( -36.60) >consensus CGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACAGCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCAACAACAGCUAUCAAGGCGUACGUACGUACAUUCACCAAGC ...((........(((((........(((((.....))))).(((((((((............)))))))))....))))).....(((.....)))((((....)))).........)) (-33.36 = -33.36 + -0.00)

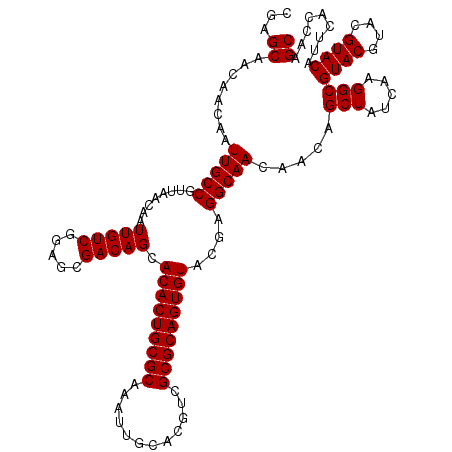
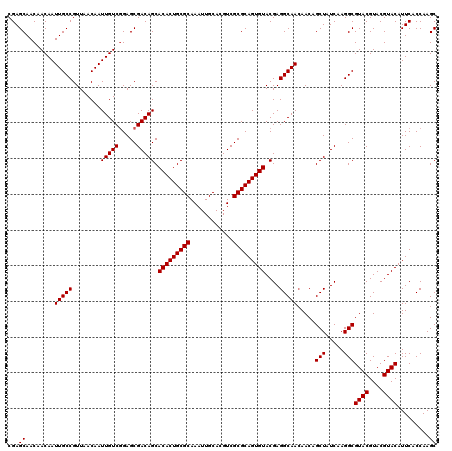
| Location | 12,076,332 – 12,076,452 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -53.86 |
| Consensus MFE | -51.26 |
| Energy contribution | -51.30 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.02 |
| Mean z-score | -4.06 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.84 |
| SVM RNA-class probability | 0.999652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076332 120 + 27905053 UGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGCUGUCGCUCUGACAAUUGUUAACGGCAAUUGUUGUUGCUCGUUGCCUGUAGCCCUGGUUCCGAUUCGGAGCGAUUAGCGCA .((((.(((((((((((((........))))))))))))).(((((((((((((((((.....))))))).......(((..(((.........)))..))).)))))))))).)).)). ( -51.60) >DroSec_CAF1 13095 120 + 1 UGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGCUGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCGUUGCCUGUAGCCCUGGUUCCGAUUCGGAGCGAUUAGCGCA .((((.(((((((((((((........))))))))))))).(((((((((((((((((.....))))))).......(((..(((.........)))..))).)))))))))).)).)). ( -53.80) >DroSim_CAF1 13316 120 + 1 UGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGCUGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCGUUGCCUGUAGCCCUGGUUCCGAUUCGGAGCGAUUAGCGCA .((((.(((((((((((((........))))))))))))).(((((((((((((((((.....))))))).......(((..(((.........)))..))).)))))))))).)).)). ( -53.80) >DroEre_CAF1 14969 120 + 1 UGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGCUGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCGUCGGCUGUAGCCCUGGUUCCGAUUCGGAGCGAUUAGCGCA .((((.(((((((((((((........))))))))))))).(((((((((((((((((.....)))))))(((..(((....(((....)))..)))..))).)))))))))).)).)). ( -54.10) >DroYak_CAF1 15237 120 + 1 UGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGCUGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCGUUGGCUGUAGCCCUGGUUCCGAUUCGGAGCGAUUAGCGCA .((((.(((((((((((((........))))))))))))).((((((((((.....((((((((((((....))))).)))))))((.(((....))).))..)))))))))).)).)). ( -56.00) >consensus UGCCUCGUACACUGCGCGACGUGCAAUUUGCGCAGUGUGCUGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCGUUGCCUGUAGCCCUGGUUCCGAUUCGGAGCGAUUAGCGCA .((((.(((((((((((((........))))))))))))).((((((((((.......((((((((((....))))).)))))..((.(((....))).))..)))))))))).)).)). (-51.26 = -51.30 + 0.04)

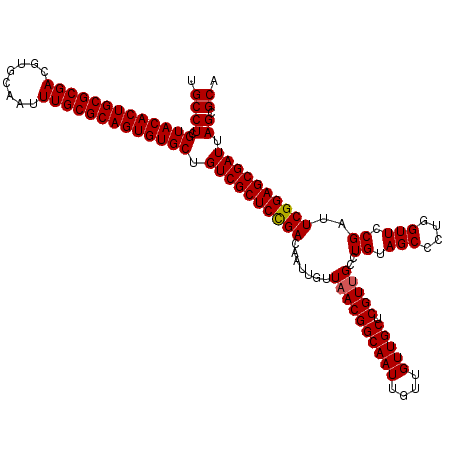
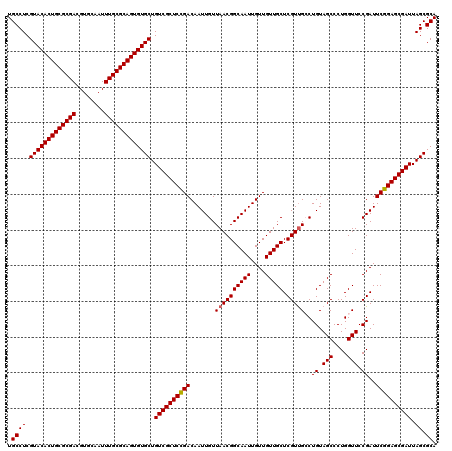
| Location | 12,076,332 – 12,076,452 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -46.32 |
| Consensus MFE | -43.64 |
| Energy contribution | -43.84 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.93 |
| SVM RNA-class probability | 0.999710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076332 120 - 27905053 UGCGCUAAUCGCUCCGAAUCGGAACCAGGGCUACAGGCAACGAGCAACAACAAUUGCCGUUAACAAUUGUCAGAGCGACAGCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCA .(((((..((((((.((............(((...(....).))).....((((((.......)))))))).)))))).)))(((((((((............))))))))).....)). ( -40.40) >DroSec_CAF1 13095 120 - 1 UGCGCUAAUCGCUCCGAAUCGGAACCAGGGCUACAGGCAACGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACAGCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCA .(((((..(((((((((............(((...(....).))).....((((((.......))))))))))))))).)))(((((((((............))))))))).....)). ( -46.50) >DroSim_CAF1 13316 120 - 1 UGCGCUAAUCGCUCCGAAUCGGAACCAGGGCUACAGGCAACGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACAGCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCA .(((((..(((((((((............(((...(....).))).....((((((.......))))))))))))))).)))(((((((((............))))))))).....)). ( -46.50) >DroEre_CAF1 14969 120 - 1 UGCGCUAAUCGCUCCGAAUCGGAACCAGGGCUACAGCCGACGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACAGCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCA .(((((..(((((((((...........(((....)))((((.((((......))))))))........))))))))).)))(((((((((............))))))))).....)). ( -49.30) >DroYak_CAF1 15237 120 - 1 UGCGCUAAUCGCUCCGAAUCGGAACCAGGGCUACAGCCAACGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACAGCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCA .(((((..(((((((((...........(((....)))((((.((((......))))))))........))))))))).)))(((((((((............))))))))).....)). ( -48.90) >consensus UGCGCUAAUCGCUCCGAAUCGGAACCAGGGCUACAGGCAACGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACAGCACACUGCGCAAAUUGCACGUCGCGCAGUGUACGAGGCA .(((((..(((((((((............(((...(....).))).....((((((.......))))))))))))))).)))(((((((((............))))))))).....)). (-43.64 = -43.84 + 0.20)

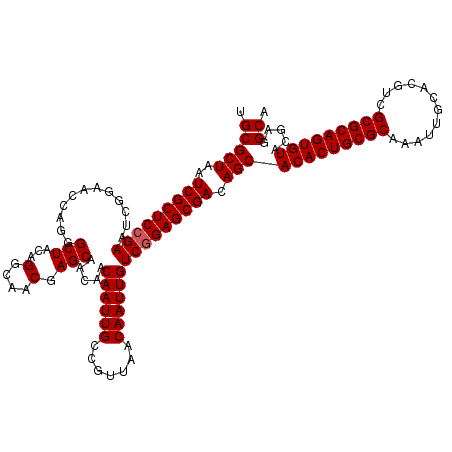
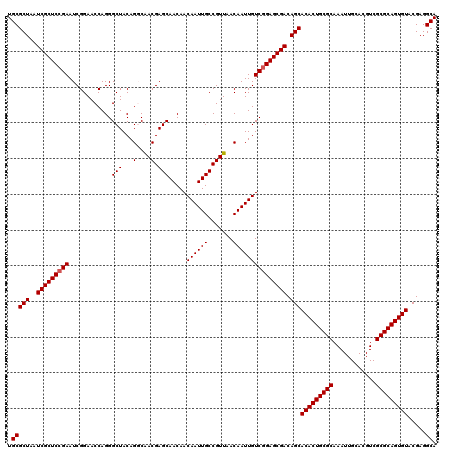
| Location | 12,076,372 – 12,076,480 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -30.66 |
| Energy contribution | -30.70 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076372 108 + 27905053 UGUCGCUCUGACAAUUGUUAACGGCAAUUGUUGUUGCUCGUUGCCUGUAGCCCUGGUUCCGAUUCGGAGCGAUUAGCGCACUUGACGCAAUGCUCUGG------------CUCUUUGGCU .(((((..((((((((((.....))))))))))..))(((..(((.........)))..))).(((((((.(((.((((....).)))))))))))))------------......))). ( -33.40) >DroSec_CAF1 13135 108 + 1 UGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCGUUGCCUGUAGCCCUGGUUCCGAUUCGGAGCGAUUAGCGCACUUGACGCAAUGCCCUGG------------CUCUUUUGCU .(((((((((((((((((.....))))))).......(((..(((.........)))..))).)))))))))).............((((.((....)------------)....)))). ( -35.70) >DroSim_CAF1 13356 108 + 1 UGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCGUUGCCUGUAGCCCUGGUUCCGAUUCGGAGCGAUUAGCGCACUUGACGCAAUGCCCUGG------------CUCUUUUGCU .(((((((((((((((((.....))))))).......(((..(((.........)))..))).)))))))))).............((((.((....)------------)....)))). ( -35.70) >DroEre_CAF1 15009 120 + 1 UGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCGUCGGCUGUAGCCCUGGUUCCGAUUCGGAGCGAUUAGCGCACUUGACGCAAUGCCCUGGCUCAACUCAACUCUCUUUUGCU .(((((((((((((((((.....)))))))(((..(((....(((....)))..)))..))).))))))))))..((((....).)))...((....))..................... ( -35.40) >DroYak_CAF1 15277 112 + 1 UGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCGUUGGCUGUAGCCCUGGUUCCGAUUCGGAGCGAUUAGCGCACUUGACGCAA--------CUCAACUCAACUCUCUUUUGCU .((((((((((.....((((((((((((....))))).)))))))((.(((....))).))..))))))))))..((((....).)))..--------...................... ( -35.40) >consensus UGUCGCUCCGACAAUUGUUAACGGCAAUUGUUGUUGCUCGUUGCCUGUAGCCCUGGUUCCGAUUCGGAGCGAUUAGCGCACUUGACGCAAUGCCCUGG____________CUCUUUUGCU .((((((((((.......((((((((((....))))).)))))..((.(((....))).))..))))))))))..((((....).)))................................ (-30.66 = -30.70 + 0.04)

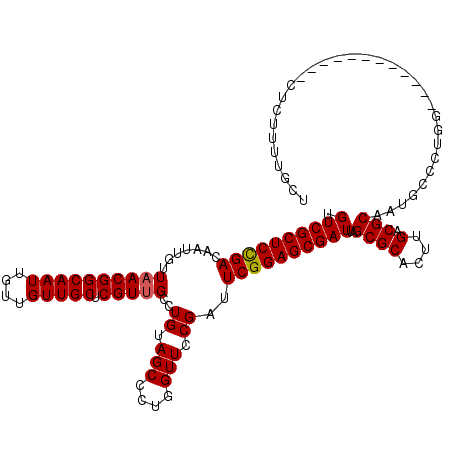
| Location | 12,076,372 – 12,076,480 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076372 108 - 27905053 AGCCAAAGAG------------CCAGAGCAUUGCGUCAAGUGCGCUAAUCGCUCCGAAUCGGAACCAGGGCUACAGGCAACGAGCAACAACAAUUGCCGUUAACAAUUGUCAGAGCGACA .(((....((------------((.(((((((((((.....)))).))).))))......(....)..))))...)))((((.((((......))))))))......((((.....)))) ( -30.60) >DroSec_CAF1 13135 108 - 1 AGCAAAAGAG------------CCAGGGCAUUGCGUCAAGUGCGCUAAUCGCUCCGAAUCGGAACCAGGGCUACAGGCAACGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACA .((......)------------).((.(((((......))))).))..(((((((((............(((...(....).))).....((((((.......))))))))))))))).. ( -34.00) >DroSim_CAF1 13356 108 - 1 AGCAAAAGAG------------CCAGGGCAUUGCGUCAAGUGCGCUAAUCGCUCCGAAUCGGAACCAGGGCUACAGGCAACGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACA .((......)------------).((.(((((......))))).))..(((((((((............(((...(....).))).....((((((.......))))))))))))))).. ( -34.00) >DroEre_CAF1 15009 120 - 1 AGCAAAAGAGAGUUGAGUUGAGCCAGGGCAUUGCGUCAAGUGCGCUAAUCGCUCCGAAUCGGAACCAGGGCUACAGCCGACGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACA .((....((.(((((.((((((((((.(((((......))))).))......((((...)))).....)))).)))).((((.((((......))))))))..))))).))...)).... ( -36.80) >DroYak_CAF1 15277 112 - 1 AGCAAAAGAGAGUUGAGUUGAG--------UUGCGUCAAGUGCGCUAAUCGCUCCGAAUCGGAACCAGGGCUACAGCCAACGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACA ................(((((.--------..((((.....))))...))(((((((...........(((....)))((((.((((......))))))))........)))))))))). ( -32.20) >consensus AGCAAAAGAG____________CCAGGGCAUUGCGUCAAGUGCGCUAAUCGCUCCGAAUCGGAACCAGGGCUACAGGCAACGAGCAACAACAAUUGCCGUUAACAAUUGUCGGAGCGACA ................................((((.....))))...(((((((((............(((...(....).))).....((((((.......))))))))))))))).. (-26.74 = -26.94 + 0.20)

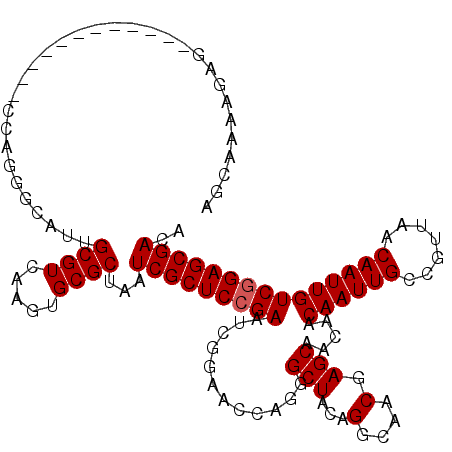
| Location | 12,076,480 – 12,076,599 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.05 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -34.92 |
| Energy contribution | -34.88 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076480 119 + 27905053 CUGUUCGCUCUACUCGUUCUCUUGUCUUAUCAGCUGGUCUGCGUGAUAAGAGCCGUACGGCAAAAAUUUUUGCAUGCAGAUCGAUGAUCAUCGGCUAUCUGCCCA-GUGGAGAAGCAGCU (((((..(((((((..............((((..(((((((((((.((((((((....)))......)))))))))))))))).))))....(((.....))).)-)))))).))))).. ( -39.90) >DroSec_CAF1 13243 119 + 1 CUGUUCGCUCUACUCGUUCUCUUGUCUUAUCAGCUGGUCUGUGUGAUAAGAGCCGUGCGGCAAAAAUUUUUGCAUGCAGAUCGAUGAUCAUCGGCUAUCUGCCCA-GUGGAGAAGAAGCU ...(((.((((((((((.((((((((....(((.....)))...))))))))....)))(((((....)))))..((((((((((....))))...))))))..)-))))))..)))... ( -35.40) >DroSim_CAF1 13464 119 + 1 CUGUUCGCUCUACUCGUUCUCUUGUCUUAUCAGCUGGUCUGCGUGAUAAGAGCCGUGCGGCAAAAAUUUUUGCAUGCAGAUCGAUGAUCAUCGGCUAUCUGCCCA-GUGGAGAAGAAGCU ...(((.(((((((..........((((((((((......)).))))))))(((((((((((((....))))).))))((((...))))..)))).........)-))))))..)))... ( -36.80) >DroEre_CAF1 15129 116 + 1 CUGCUCGCUCUACUCGUUCUCUUGUCUUAUCAGCUGGUCUGCGUGAUAAGA-CCGCAAGGCAGAAAUUUUUGCAUGCAGAUCGAUGAUCAUCGGCUAUCUGCCCAAGUGGAGA---AGCU ..(((..(((((((.........(((((((((((......)).))))))))-).....((((((....((((....))))(((((....)))))...))))))..))))))).---))). ( -39.80) >DroYak_CAF1 15389 117 + 1 CUGGUCGCUCUACUCGUGCUCUUGUCUUAUCAGCUGGUCUGCGUGAUAAGAGCCGCACGGCAGAAAUUUUUGCAUGCAGAUCGAUGAUCAUCGGCUAUCUGCCCAAGUGGGGA---AGCU ..(((..((((((((((((.....((((((((((......)).))))))))...)))))(((((....)))))..((((((((((....))))...))))))...))))))).---.))) ( -41.50) >consensus CUGUUCGCUCUACUCGUUCUCUUGUCUUAUCAGCUGGUCUGCGUGAUAAGAGCCGUACGGCAAAAAUUUUUGCAUGCAGAUCGAUGAUCAUCGGCUAUCUGCCCA_GUGGAGAAG_AGCU ..(((..((((((...............((((..(((((((((((.((((((((....)))......)))))))))))))))).))))....(((.....)))...))))))....))). (-34.92 = -34.88 + -0.04)

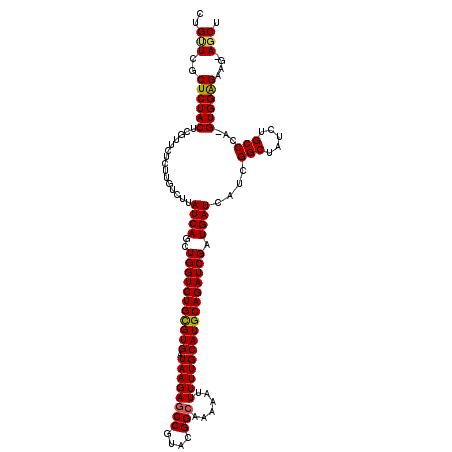
| Location | 12,076,480 – 12,076,599 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.05 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -28.22 |
| Energy contribution | -28.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12076480 119 - 27905053 AGCUGCUUCUCCAC-UGGGCAGAUAGCCGAUGAUCAUCGAUCUGCAUGCAAAAAUUUUUGCCGUACGGCUCUUAUCACGCAGACCAGCUGAUAAGACAAGAGAACGAGUAGAGCGAACAG ..(((.(((((.((-(.(((((((...((((....)))))))))).........((((((((....)))((((((((.((......))))))))))..))))).).))).))).)).))) ( -33.60) >DroSec_CAF1 13243 119 - 1 AGCUUCUUCUCCAC-UGGGCAGAUAGCCGAUGAUCAUCGAUCUGCAUGCAAAAAUUUUUGCCGCACGGCUCUUAUCACACAGACCAGCUGAUAAGACAAGAGAACGAGUAGAGCGAACAG .((((.(((((..(-((.((((((...((((....))))))))))..(((((....)))))....))).(((((((((........).))))))))...))))).))))........... ( -30.60) >DroSim_CAF1 13464 119 - 1 AGCUUCUUCUCCAC-UGGGCAGAUAGCCGAUGAUCAUCGAUCUGCAUGCAAAAAUUUUUGCCGCACGGCUCUUAUCACGCAGACCAGCUGAUAAGACAAGAGAACGAGUAGAGCGAACAG .((((.(((((..(-((.((((((...((((....))))))))))..(((((....)))))....))).((((((((.((......))))))))))...))))).))))........... ( -34.10) >DroEre_CAF1 15129 116 - 1 AGCU---UCUCCACUUGGGCAGAUAGCCGAUGAUCAUCGAUCUGCAUGCAAAAAUUUCUGCCUUGCGG-UCUUAUCACGCAGACCAGCUGAUAAGACAAGAGAACGAGUAGAGCGAGCAG .(((---((((.(((((.((((((...((((....))))))))))..(((........)))(((...(-((((((((.((......)))))))))))..)))..))))).))).)))).. ( -41.70) >DroYak_CAF1 15389 117 - 1 AGCU---UCCCCACUUGGGCAGAUAGCCGAUGAUCAUCGAUCUGCAUGCAAAAAUUUCUGCCGUGCGGCUCUUAUCACGCAGACCAGCUGAUAAGACAAGAGCACGAGUAGAGCGACCAG ....---........(((((((((...((((....))))))))))..........(((((((((((.(.((((((((.((......)))))))))))....))))).))))))...))). ( -39.50) >consensus AGCU_CUUCUCCAC_UGGGCAGAUAGCCGAUGAUCAUCGAUCUGCAUGCAAAAAUUUUUGCCGUACGGCUCUUAUCACGCAGACCAGCUGAUAAGACAAGAGAACGAGUAGAGCGAACAG .(((....(((.......((((((...((((....)))))))))).........((((((((....)))((((((((.((......))))))))))..)))))..)))...)))...... (-28.22 = -28.78 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:26 2006