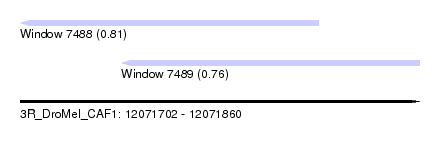
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,071,702 – 12,071,860 |
| Length | 158 |
| Max. P | 0.813156 |

| Location | 12,071,702 – 12,071,820 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -20.88 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12071702 118 - 27905053 ACUCACACACUCAUGCAAAUAUAGCAGCG-GCUAUGUGGCUAUAUGUGCAAC-CAAAGCGUAACCAAAACCAAACCAACCAAGUCGCGAUUUUCAUUCAUAAAAUCGAAGAGCCAAAGAA .........(((.((((.(((((((.(((-....))).))))))).))))..-.................................(((((((.......)))))))..)))........ ( -21.60) >DroSec_CAF1 8235 120 - 1 ACUCACACACUCAUGCAAAUAUAGCAGCGGGCUAUGUGGCUAUAUGUGCAACCCAAAGCGUAACCAAAACCAAACCAACCAAGUCGCGAUUUUCAUUCAUAAAAUCGAAGAGCCAAAGAA .........(((.((((.(((((((.(((.....))).))))))).))))....................................(((((((.......)))))))..)))........ ( -21.60) >DroSim_CAF1 8670 118 - 1 ACUCACACACUCAUGCAAAUAUAGCAGCG-GCUAUGUGGCUAUAUGUGCAAC-CAAAGCGUAACCAAAACCAAACCAACCAAGUCGCGAUUUUCAUUCAUAAAAUCGAAGAGCCAAAGAA .........(((.((((.(((((((.(((-....))).))))))).))))..-.................................(((((((.......)))))))..)))........ ( -21.60) >DroEre_CAF1 10227 118 - 1 ACUCACACACUCAUGCAAAUAUAGCAGCG-GCUAUGUGGCUAUAUGUGCAAC-CAAAGCGUAACCAAAACCAAACCAACCAAGUCGCGAUUUUCAUUCAUAAAAUCGAAGAGCCAAAGAA .........(((.((((.(((((((.(((-....))).))))))).))))..-.................................(((((((.......)))))))..)))........ ( -21.60) >DroYak_CAF1 10432 118 - 1 ACUCACACACUCAUGCAAAUAUAGCAGCG-GCUAUGUGGCUAUAUGUGCAAC-CAAAGCGUAACCAAAACCAAACCAACCAAGUCGCGAUUUUCAUUCAUAAAAUCGAAGAGCCAAAGAA .........(((.((((.(((((((.(((-....))).))))))).))))..-.................................(((((((.......)))))))..)))........ ( -21.60) >consensus ACUCACACACUCAUGCAAAUAUAGCAGCG_GCUAUGUGGCUAUAUGUGCAAC_CAAAGCGUAACCAAAACCAAACCAACCAAGUCGCGAUUUUCAUUCAUAAAAUCGAAGAGCCAAAGAA .........(((.((((.(((((((.(((.....))).))))))).))))....................................(((((((.......)))))))..)))........ (-20.88 = -20.88 + 0.00)

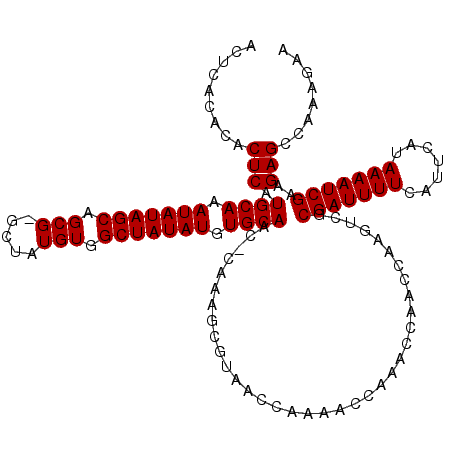
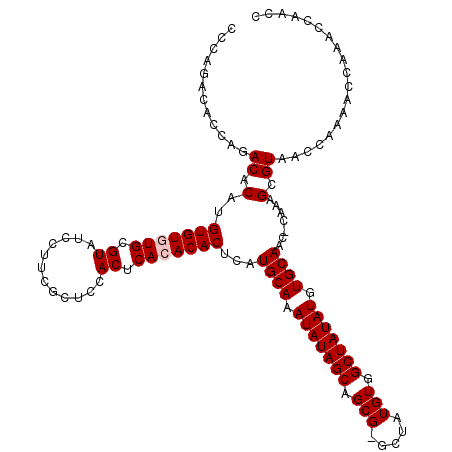
| Location | 12,071,742 – 12,071,860 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.82 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.86 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12071742 118 - 27905053 CCCAGACACCAGACACAUGUGUGUGCGUAUCCUUCGCUCCACUCACACACUCAUGCAAAUAUAGCAGCG-GCUAUGUGGCUAUAUGUGCAAC-CAAAGCGUAACCAAAACCAAACCAACC ............((.(..(((((((.((............)).)))))))...((((.(((((((.(((-....))).))))))).))))..-....).))................... ( -25.30) >DroSec_CAF1 8275 120 - 1 CCCAGACACCAGACACAUGUGUAUGCGUAUCCUUCGCUCCACUCACACACUCAUGCAAAUAUAGCAGCGGGCUAUGUGGCUAUAUGUGCAACCCAAAGCGUAACCAAAACCAAACCAACC ............((.(.((((...(((.......)))......))))......((((.(((((((.(((.....))).))))))).)))).......).))................... ( -21.00) >DroSim_CAF1 8710 118 - 1 CCCAGACACCAGACACAUGUGUAUGCGUAUCCUUCGCUCCACUCACACACUCAUGCAAAUAUAGCAGCG-GCUAUGUGGCUAUAUGUGCAAC-CAAAGCGUAACCAAAACCAAACCAACC ............((.(.((((...(((.......)))......))))......((((.(((((((.(((-....))).))))))).))))..-....).))................... ( -21.00) >DroEre_CAF1 10267 118 - 1 CCCAGACACCAGACACAUGUGUGUGCGUAUCCUUCGCUCCACUCACACACUCAUGCAAAUAUAGCAGCG-GCUAUGUGGCUAUAUGUGCAAC-CAAAGCGUAACCAAAACCAAACCAACC ............((.(..(((((((.((............)).)))))))...((((.(((((((.(((-....))).))))))).))))..-....).))................... ( -25.30) >DroYak_CAF1 10472 118 - 1 CCCAGACACCAGACACAUGUGUGUGCGUAUCCUUCGCUCCACUCACACACUCAUGCAAAUAUAGCAGCG-GCUAUGUGGCUAUAUGUGCAAC-CAAAGCGUAACCAAAACCAAACCAACC ............((.(..(((((((.((............)).)))))))...((((.(((((((.(((-....))).))))))).))))..-....).))................... ( -25.30) >consensus CCCAGACACCAGACACAUGUGUGUGCGUAUCCUUCGCUCCACUCACACACUCAUGCAAAUAUAGCAGCG_GCUAUGUGGCUAUAUGUGCAAC_CAAAGCGUAACCAAAACCAAACCAACC ............((.(..(((((((.((............)).)))))))...((((.(((((((.(((.....))).))))))).)))).......).))................... (-22.46 = -22.86 + 0.40)

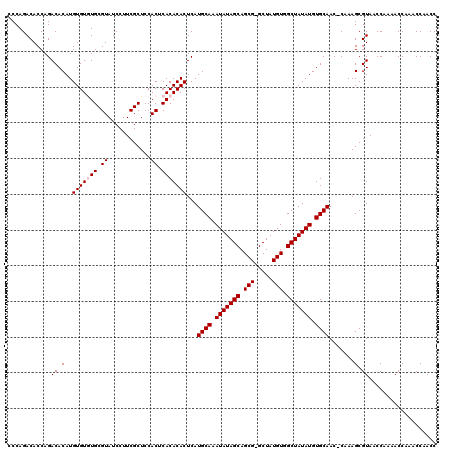
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:03 2006