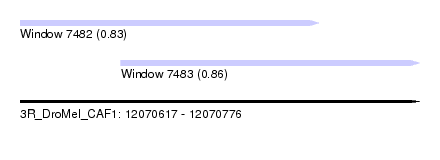
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,070,617 – 12,070,776 |
| Length | 159 |
| Max. P | 0.859896 |

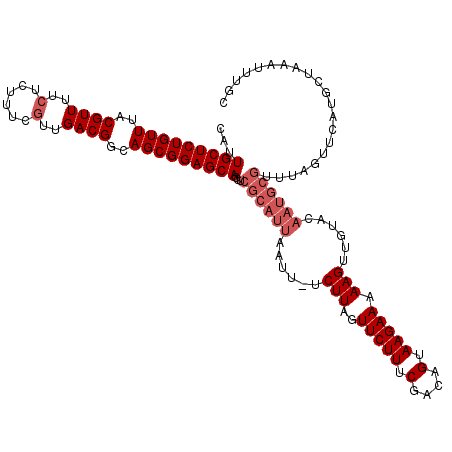
| Location | 12,070,617 – 12,070,736 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -26.62 |
| Energy contribution | -27.02 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12070617 119 + 27905053 ACAAACUUUUUGGCGUCGCCGUCGUCGUCAGCGUCAGCGUCAUUGCUCUGUUUACGUUUUCUCUUUCGUUGACGGCAGCGGAGCAGCUCGCAUUAAUU-UCUUAGUUCUUUCGACAGUAA ....(((..((((((.((....)).)))))).(((.(((...((((((((((..((((..(......)..))))..))))))))))..)))(((((..-..)))))......)))))).. ( -33.30) >DroSec_CAF1 7157 113 + 1 ACAAACUUUUUGGCGUCGCCGUCG------GCGUCAGCGUCAUUGCUCUGUUUACGUUUUCUCUUUCGUUGACGGCAGCGGAGCAGCUCGCAUUAAUU-UCUUAGUUCUUUCGACAGUAA ...((((...((((((((....))------))))))(((...((((((((((..((((..(......)..))))..))))))))))..))).......-....))))............. ( -33.70) >DroSim_CAF1 7593 113 + 1 ACAAACUUUUUGGCGUCGCCGUCG------GCGUCAGCGUCAUUGCUCUGUUUACGUUUUCUCUUUCGUUGACGGCAGCGGAGCAGCUCGCAUUAAUU-UCUUAGUUCUUUCUACAGUAA ...((((...((((((((....))------))))))(((...((((((((((..((((..(......)..))))..))))))))))..))).......-....))))............. ( -33.70) >DroEre_CAF1 9183 114 + 1 ACAAACUUUUUGGCGUCGCUGUCC------GCGUCAGCGUCAUUGCUCUGUUUACGUUUUCUCUUUCGUUGACGGCAGCGGAGCACCUCGCAUUAAUUUUCUUAGUUCUUUCGACAGUAA .................((((((.------......(((....(((((((((..((((..(......)..))))..)))))))))...)))(((((.....)))))......)))))).. ( -29.20) >DroYak_CAF1 9275 113 + 1 ACAAACUUUUUGGCGUCGCCGUCG------GCGUCAGCGUCAUUGCUCUGUUUACGUUUUCUCUUUCCUUGACGGCAGCGGAGCAGUUCGCAUUAAUU-UCUUAGUUCUUUCGACAGUAA ...((((...((((((((....))------))))))(((..(((((((((((..((((............))))..))))))))))).))).......-....))))............. ( -33.30) >consensus ACAAACUUUUUGGCGUCGCCGUCG______GCGUCAGCGUCAUUGCUCUGUUUACGUUUUCUCUUUCGUUGACGGCAGCGGAGCAGCUCGCAUUAAUU_UCUUAGUUCUUUCGACAGUAA ...((((...((((((..............))))))(((...((((((((((..((((..(......)..))))..))))))))))..)))............))))............. (-26.62 = -27.02 + 0.40)

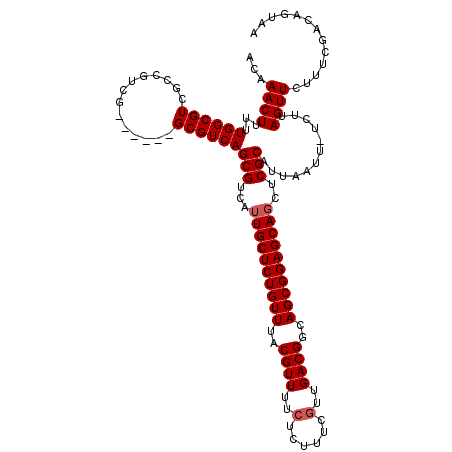
| Location | 12,070,657 – 12,070,776 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.35 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -22.30 |
| Energy contribution | -23.70 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12070657 119 + 27905053 CAUUGCUCUGUUUACGUUUUCUCUUUCGUUGACGGCAGCGGAGCAGCUCGCAUUAAUU-UCUUAGUUCUUUCGACAGUAAGAAAAAGUUGUACUAUGCGUUUAGUUCAUGUUGAAUUUGC ..((((((((((..((((..(......)..))))..))))))))))..(((((..((.-.(((..(((((.(....).))))).)))..))...)))))...(((((.....)))))... ( -28.80) >DroSec_CAF1 7191 119 + 1 CAUUGCUCUGUUUACGUUUUCUCUUUCGUUGACGGCAGCGGAGCAGCUCGCAUUAAUU-UCUUAGUUCUUUCGACAGUAAGAAAAAGUUUUACAAUGCGUUUAGUUCAUGCUAAAUUUGC ..((((((((((..((((..(......)..))))..))))))))))..((((((..((-(((((.(((....)).).)))))))..(.....)))))))((((((....))))))..... ( -30.70) >DroSim_CAF1 7627 119 + 1 CAUUGCUCUGUUUACGUUUUCUCUUUCGUUGACGGCAGCGGAGCAGCUCGCAUUAAUU-UCUUAGUUCUUUCUACAGUAAGAAAAAGUUUUACAAUGCGUUUAGUUCAUGCUAAAUUUGC ..((((((((((..((((..(......)..))))..))))))))))..((((((....-...(((......)))..((((((.....))))))))))))((((((....))))))..... ( -30.30) >DroEre_CAF1 9217 95 + 1 CAUUGCUCUGUUUACGUUUUCUCUUUCGUUGACGGCAGCGGAGCACCUCGCAUUAAUUUUCUUAGUUCUUUCGACAGUAAGAAAAAGUUGUUCAC------------------------- ...(((((((((..((((..(......)..))))..)))))))))....(((....((((((((.(((....)).).))))))))...)))....------------------------- ( -23.50) >DroYak_CAF1 9309 119 + 1 CAUUGCUCUGUUUACGUUUUCUCUUUCCUUGACGGCAGCGGAGCAGUUCGCAUUAAUU-UCUUAGUUCUUUCGACAGUAAGAAAAAGUUGUACAAUGCGUUGAGUUCAUGCUGAAUUUUC .(((((((((((..((((............))))..))))))))))).((((((.((.-.(((..(((((.(....).))))).)))..))..))))))..((((((.....)))))).. ( -29.70) >consensus CAUUGCUCUGUUUACGUUUUCUCUUUCGUUGACGGCAGCGGAGCAGCUCGCAUUAAUU_UCUUAGUUCUUUCGACAGUAAGAAAAAGUUGUACAAUGCGUUUAGUUCAUGCUAAAUUUGC ...(((((((((..((((..(......)..))))..)))))))))...((((((......(((..(((((.(....).))))).)))......))))))..................... (-22.30 = -23.70 + 1.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:57 2006