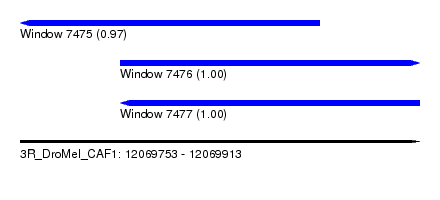
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,069,753 – 12,069,913 |
| Length | 160 |
| Max. P | 0.999907 |

| Location | 12,069,753 – 12,069,873 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -34.49 |
| Energy contribution | -34.13 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12069753 120 - 27905053 AUUGCUGAAUGGCAGCAGCCACCUAGAGUGGGCAUUGUUGCUGUUCUUGGGUUCGCUGCCAUGUCCCUUUGUUGUUGAGAACCUUCAUUACUCCUUACCCCAUGGUUUUGACUGGUAUUA ..((((..((((((((((((....((((..(.((....)))..))))..)))).))))))))...........(((.((((((....................))))))))).))))... ( -32.85) >DroSec_CAF1 6365 120 - 1 AUUGCUGAAUGGCAGCAGCCACCGAGAGCGGGCAUUGUUGCUGUUCUUGGGUUCGCUGCCAUGUCCCUUUGUUGCUGAGAACCUUCAUUACUCCUUACCCCAUGGUUUUGGCUGGCAUUA ..((((..((((((((((((..(((((((((.((....))))))))))))))).))))))))...........(((.((((((....................))))))))).))))... ( -41.55) >DroSim_CAF1 6813 120 - 1 AUUGCUGAAUGGCAGCAGCCACCGAGAGCGGGCAUUGUUGCUGUUCUUGGGUUCGCUGCCAUGUCCCUUUGUUGCUGAGAACCUUCAUUACUCCUUACCCCAUGGCUUUGGCUGGUAUUA .(((((....)))))((((((((((((((((.((....)))))))))))).......((((((.......((...((((....))))..)).........))))))..))))))...... ( -38.89) >DroEre_CAF1 8287 117 - 1 AUUGCUGAAUGGCAGCAGCCACCGAGAGCGAGCAUUGUUGCUGUUCUUG-GUUCGCUGCCAUGUCCCUUUGUUGUUGGGAACCUUCAUUACUCCUUAUCCCUUGCUUUUGGCUGGUAA-- ...((..((.(((((((((.(((((((((.((((....)))))))))))-))..)))))....((((.........))))......................))))))..))......-- ( -40.90) >DroYak_CAF1 8414 118 - 1 AUUGCUGAAUGGCAGCAGCCACCGAGAGCGGGCAUUGUUGCUGUUCUUGGGUUCGCUGUCAUGUCCCUUUGUUGCCGAGAACCUUCAUUACUCCUUAUCCCUUGGAUUUGGCUGGUAU-- ........((((((((((((..(((((((((.((....))))))))))))))).))))))))..........(((((((....)))......((..((((...))))..))..)))).-- ( -36.90) >consensus AUUGCUGAAUGGCAGCAGCCACCGAGAGCGGGCAUUGUUGCUGUUCUUGGGUUCGCUGCCAUGUCCCUUUGUUGCUGAGAACCUUCAUUACUCCUUACCCCAUGGUUUUGGCUGGUAUUA ..((((..((((((((((((..(((((((.((((....))))))))))))))).))))))))...........(((.((((((....................))))))))).))))... (-34.49 = -34.13 + -0.36)

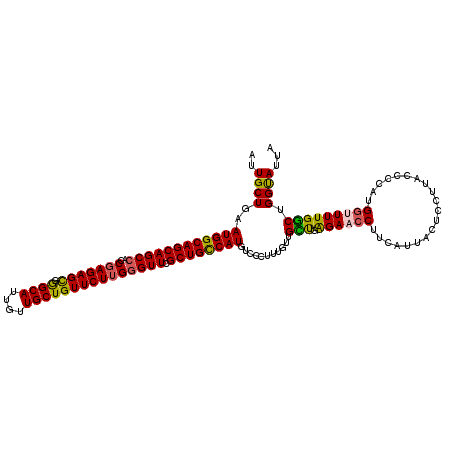
| Location | 12,069,793 – 12,069,913 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -37.48 |
| Energy contribution | -38.28 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.55 |
| SVM RNA-class probability | 0.999372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12069793 120 + 27905053 UCUCAACAACAAAGGGACAUGGCAGCGAACCCAAGAACAGCAACAAUGCCCACUCUAGGUGGCUGCUGCCAUUCAGCAAUGUAAACAAUGCACAAGUUGCACGUGCAGCCAGCAGGCUGG ......((((.....((.(((((((((..(((.(((...(((....)))....))).)).)..))))))))))).(((.((....)).)))....))))......(((((....))))). ( -38.90) >DroSec_CAF1 6405 120 + 1 UCUCAGCAACAAAGGGACAUGGCAGCGAACCCAAGAACAGCAACAAUGCCCGCUCUCGGUGGCUGCUGCCAUUCAGCAAUGUAAACAAUGCACAAGUUGCACGUGCAGCCAGCAGGCUGG .....(((((.....((.(((((((((..(((.(((.(.(((....)))..).))).)).)..))))))))))).(((.((....)).)))....))))).....(((((....))))). ( -43.90) >DroSim_CAF1 6853 120 + 1 UCUCAGCAACAAAGGGACAUGGCAGCGAACCCAAGAACAGCAACAAUGCCCGCUCUCGGUGGCUGCUGCCAUUCAGCAAUGUAAACAAUGCACAAGUUGCACGUGCAGCCAGCAGGCUGG .....(((((.....((.(((((((((..(((.(((.(.(((....)))..).))).)).)..))))))))))).(((.((....)).)))....))))).....(((((....))))). ( -43.90) >DroEre_CAF1 8325 119 + 1 UCCCAACAACAAAGGGACAUGGCAGCGAAC-CAAGAACAGCAACAAUGCUCGCUCUCGGUGGCUGCUGCCAUUCAGCAAUGUAAACAAUGCACAAGUUGCACGUGCAGCCAGCAGGCUGG ((((.........))))(((((((((((((-(.(((.(((((....)))).).))).)))(((....)))..)).(((.((....)).)))....))))).))))(((((....))))). ( -41.70) >DroYak_CAF1 8452 120 + 1 UCUCGGCAACAAAGGGACAUGACAGCGAACCCAAGAACAGCAACAAUGCCCGCUCUCGGUGGCUGCUGCCAUUCAGCAAUGUAAACAAUGCACAAGUUGCACGUGCAGCCAGCAGGCUGG .(.(((((((.....((.(((.(((((..(((.(((.(.(((....)))..).))).)).)..))))).))))).(((.((....)).)))....))))).)).)(((((....))))). ( -37.60) >consensus UCUCAGCAACAAAGGGACAUGGCAGCGAACCCAAGAACAGCAACAAUGCCCGCUCUCGGUGGCUGCUGCCAUUCAGCAAUGUAAACAAUGCACAAGUUGCACGUGCAGCCAGCAGGCUGG .....(((((.....((.(((((((((..(((.(((...(((....)))....))).)).)..))))))))))).(((.((....)).)))....))))).....(((((....))))). (-37.48 = -38.28 + 0.80)

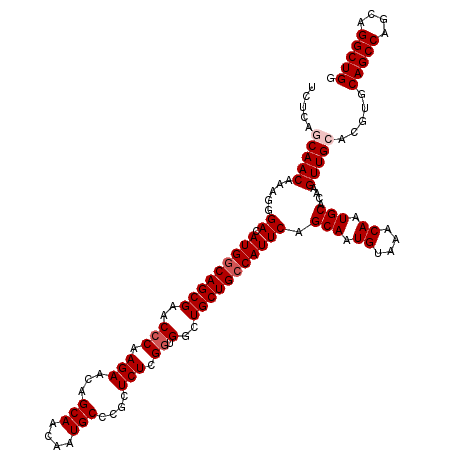
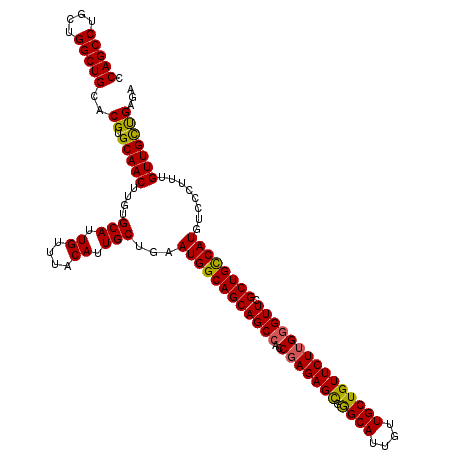
| Location | 12,069,793 – 12,069,913 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -47.84 |
| Consensus MFE | -46.46 |
| Energy contribution | -45.98 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.48 |
| SVM RNA-class probability | 0.999907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12069793 120 - 27905053 CCAGCCUGCUGGCUGCACGUGCAACUUGUGCAUUGUUUACAUUGCUGAAUGGCAGCAGCCACCUAGAGUGGGCAUUGUUGCUGUUCUUGGGUUCGCUGCCAUGUCCCUUUGUUGUUGAGA .(((((....)))))..(..(((((....(((.((....)).)))...((((((((((((....((((..(.((....)))..))))..)))).))))))))........)))))...). ( -42.80) >DroSec_CAF1 6405 120 - 1 CCAGCCUGCUGGCUGCACGUGCAACUUGUGCAUUGUUUACAUUGCUGAAUGGCAGCAGCCACCGAGAGCGGGCAUUGUUGCUGUUCUUGGGUUCGCUGCCAUGUCCCUUUGUUGCUGAGA .(((((....)))))..((.(((((....(((.((....)).)))...((((((((((((..(((((((((.((....))))))))))))))).))))))))........)))))))... ( -49.30) >DroSim_CAF1 6853 120 - 1 CCAGCCUGCUGGCUGCACGUGCAACUUGUGCAUUGUUUACAUUGCUGAAUGGCAGCAGCCACCGAGAGCGGGCAUUGUUGCUGUUCUUGGGUUCGCUGCCAUGUCCCUUUGUUGCUGAGA .(((((....)))))..((.(((((....(((.((....)).)))...((((((((((((..(((((((((.((....))))))))))))))).))))))))........)))))))... ( -49.30) >DroEre_CAF1 8325 119 - 1 CCAGCCUGCUGGCUGCACGUGCAACUUGUGCAUUGUUUACAUUGCUGAAUGGCAGCAGCCACCGAGAGCGAGCAUUGUUGCUGUUCUUG-GUUCGCUGCCAUGUCCCUUUGUUGUUGGGA .((((.((..(((((((((.......))))))..)))..))..)))).((((((((....(((((((((.((((....)))))))))))-))..)))))))).((((.........)))) ( -49.00) >DroYak_CAF1 8452 120 - 1 CCAGCCUGCUGGCUGCACGUGCAACUUGUGCAUUGUUUACAUUGCUGAAUGGCAGCAGCCACCGAGAGCGGGCAUUGUUGCUGUUCUUGGGUUCGCUGUCAUGUCCCUUUGUUGCCGAGA .(((((....)))))..((.(((((....(((.((....)).)))...((((((((((((..(((((((((.((....))))))))))))))).))))))))........)))))))... ( -48.80) >consensus CCAGCCUGCUGGCUGCACGUGCAACUUGUGCAUUGUUUACAUUGCUGAAUGGCAGCAGCCACCGAGAGCGGGCAUUGUUGCUGUUCUUGGGUUCGCUGCCAUGUCCCUUUGUUGCUGAGA .(((((....)))))..((.(((((....(((.((....)).)))...((((((((((((..(((((((.((((....))))))))))))))).))))))))........)))))))... (-46.46 = -45.98 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:51 2006