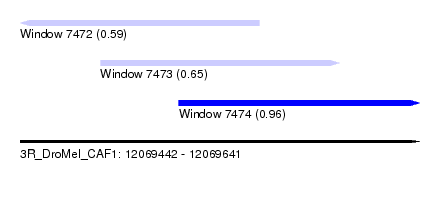
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,069,442 – 12,069,641 |
| Length | 199 |
| Max. P | 0.963219 |

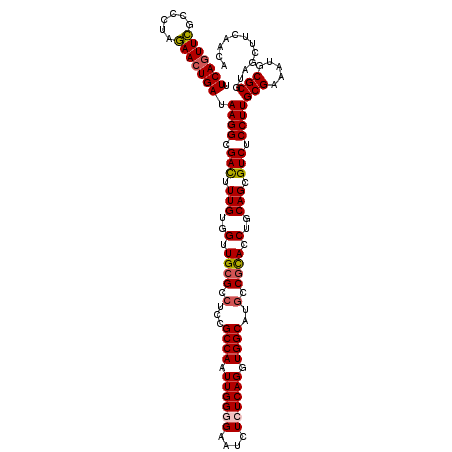
| Location | 12,069,442 – 12,069,561 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.39 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -24.03 |
| Energy contribution | -23.78 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12069442 119 - 27905053 CAACCACAAAGUCGCCUUAUCAGUUCUAGGGCGAACUGAACUUGUGGCAAGUUCAAGUUACUG-UAAGUCAGUUCGAGGGAUUGAUAUUACUUGGAAUUUGAGCCAGGCACUAAUCUUAC ....(((((((((((((((.......))))))).)))....)))))((..(((((((((...(-((((((((((.....))))))).))))....)))))))))...))........... ( -34.90) >DroSec_CAF1 6064 104 - 1 CAACCACAAAGUCGCCUUAUCAGUUCUAGGGCGAACUGAACUUGUGGCAAGUUC----------------AGUUUGAGGGAUUGGUAUUACUUGGAAUUUGAGCCAGGCAAUAAUCUUAC .............(((.((((((((((....(((((((((((((...)))))))----------------)))))).)))))))))).....(((........))))))........... ( -32.60) >DroSim_CAF1 6496 120 - 1 CAACCACAAAGUCGCCUUAUCAGUUCUAGGGCGAACUGAACUUGUGGCAAGUUCAAGUUACUGAUAAGUCAGUUUGAGGGAUUGGUAUUACUUGGAAUUUGAGCCAGGCAAUAAUCUUAC ....(((((((((((((((.......))))))).)))....)))))((..((((((((((((((....)))))(..((..((....))..))..))))))))))...))........... ( -32.30) >DroEre_CAF1 7978 118 - 1 CAACCACAAAAUCGCCUUAUCAGUUUUAG-ACAAAUUGAACUUGUGGCAAGUUCAACUUACUGAUA-GUCAGUUUGAGGGAUUGGUACUACUUGGAAUUUGAGCCAUGCGCUUAUCUCAC ......((((...((..(((((((...((-.....(((((((((...))))))))))).)))))))-))...)))).(((((.((..(....(((........))).)..)).))))).. ( -26.20) >DroYak_CAF1 8101 119 - 1 CAACCACAACAUCGCCUUAUCAGUUCUAG-ACGAACUGAACUUGUGGCAAGUUCAAGUUACUGAUAAGUCAGUUUGAGGGAUUGGUAUUACUUGGAAUGUGAGCCAGGCGCUAAUCUCGC .............(.(((((((((....(-((....((((((((...)))))))).)))))))))))).).......(((((((((....(((((........))))).))))))))).. ( -34.10) >consensus CAACCACAAAGUCGCCUUAUCAGUUCUAGGGCGAACUGAACUUGUGGCAAGUUCAAGUUACUGAUAAGUCAGUUUGAGGGAUUGGUAUUACUUGGAAUUUGAGCCAGGCACUAAUCUUAC .............(((.((((((((((....(((((((((((((...))))))).....(((....))).)))))).)))))))))).....(((........))))))........... (-24.03 = -23.78 + -0.25)



| Location | 12,069,482 – 12,069,601 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -27.83 |
| Energy contribution | -28.08 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12069482 119 + 27905053 CCCUCGAACUGACUUA-CAGUAACUUGAACUUGCCACAAGUUCAGUUCGCCCUAGAACUGAUAAGGCGACUUUGUGGUUGAGCCUCCGCCAAUUGGGGAAUCUCUCAGGUGGCAUGCCGC .......((((.....-)))).((((((....(((((((..(((((((......)))))))..((....))))))))).(((..(((.((....)))))..)))))))))((....)).. ( -36.80) >DroSec_CAF1 6104 104 + 1 CCCUCAAACU----------------GAACUUGCCACAAGUUCAGUUCGCCCUAGAACUGAUAAGGCGACUUUGUGGUUGCGCCUCUGCCAAUUGGGGAAUCUCUCAGGUGGCAUGCCGC .((.((((((----------------(((((((...))))))))))(((((.((.......)).)))))..))).))..(((.(..(((((.((((((....)))))).))))).).))) ( -36.50) >DroSim_CAF1 6536 120 + 1 CCCUCAAACUGACUUAUCAGUAACUUGAACUUGCCACAAGUUCAGUUCGCCCUAGAACUGAUAAGGCGACUUUGUGGUUGCGCCUCUGCCAAUUGGGGAAUCUCUCAGGUGGCAUGCCGC ............(((((((((...(((((((((...))))))))).((......)))))))))))((((((....))))))((...(((((.((((((....)))))).))))).))... ( -39.70) >DroEre_CAF1 8018 118 + 1 CCCUCAAACUGAC-UAUCAGUAAGUUGAACUUGCCACAAGUUCAAUUUGU-CUAAAACUGAUAAGGCGAUUUUGUGGUUGCGCCUCCGCCAAUUGGUGAAUCUCUCAGGUGGCAUACCGU .............-(((((((((((((((((((...))))))))))))..-.....)))))))..((((((....))))))(((.(((((....)))((.....)).)).)))....... ( -32.81) >DroYak_CAF1 8141 119 + 1 CCCUCAAACUGACUUAUCAGUAACUUGAACUUGCCACAAGUUCAGUUCGU-CUAGAACUGAUAAGGCGAUGUUGUGGUUGCGCCUCCGCCAAUUGGUGAAUCUCUCAGGUGGCAUGCCGC ........((((.......((((((..((((((((......(((((((..-...)))))))...))))).)))..)))))).....((((....))))......))))((((....)))) ( -36.60) >consensus CCCUCAAACUGACUUAUCAGUAACUUGAACUUGCCACAAGUUCAGUUCGCCCUAGAACUGAUAAGGCGACUUUGUGGUUGCGCCUCCGCCAAUUGGGGAAUCUCUCAGGUGGCAUGCCGC ...((((((((......))))...))))..(((((......(((((((......)))))))...)))))....(((((...(((.((.((....)).((.....)).)).)))..))))) (-27.83 = -28.08 + 0.25)

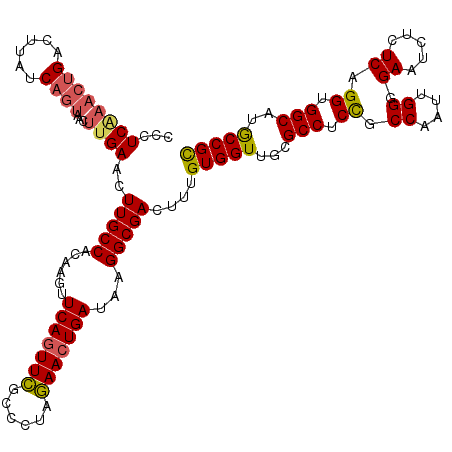
| Location | 12,069,521 – 12,069,641 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.16 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -38.82 |
| Energy contribution | -39.10 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12069521 120 + 27905053 UUCAGUUCGCCCUAGAACUGAUAAGGCGACUUUGUGGUUGAGCCUCCGCCAAUUGGGGAAUCUCUCAGGUGGCAUGCCGCACCUGCAGCGUCUCCUUGCGAAAUGCGCGUAGCUUCAACA .(((((((......))))))).((((.(((.(((((((.(((..(((.((....)))))..)))....((((....))))))).)))).))).))))(((.....)))............ ( -40.50) >DroSec_CAF1 6128 120 + 1 UUCAGUUCGCCCUAGAACUGAUAAGGCGACUUUGUGGUUGCGCCUCUGCCAAUUGGGGAAUCUCUCAGGUGGCAUGCCGCACCUGCAGCGUCUCCUUGCGAAAUGCGCGUAGCUUCAACA .(((((((......))))))).((((.(((.(((..(.((((.(..(((((.((((((....)))))).))))).).)))).)..))).))).))))(((.....)))............ ( -45.50) >DroSim_CAF1 6576 120 + 1 UUCAGUUCGCCCUAGAACUGAUAAGGCGACUUUGUGGUUGCGCCUCUGCCAAUUGGGGAAUCUCUCAGGUGGCAUGCCGCACCUGCAGCGUCUCCUUGCGAAAUGCGCGUAGCUUCAACA .(((((((......))))))).((((.(((.(((..(.((((.(..(((((.((((((....)))))).))))).).)))).)..))).))).))))(((.....)))............ ( -45.50) >DroEre_CAF1 8057 119 + 1 UUCAAUUUGU-CUAAAACUGAUAAGGCGAUUUUGUGGUUGCGCCUCCGCCAAUUGGUGAAUCUCUCAGGUGGCAUACCGUACCUGCAGCGUCUCCUUGCGAAAUGCGCGUAGCUUCAACA .....(((((-(.......))))))((((((((((((..((((...((((....)))).......((((((........))))))..))))..))..))))))).)))............ ( -31.80) >DroYak_CAF1 8181 119 + 1 UUCAGUUCGU-CUAGAACUGAUAAGGCGAUGUUGUGGUUGCGCCUCCGCCAAUUGGUGAAUCUCUCAGGUGGCAUGCCGCACCUGCAGCGUCUCCUUGCGAAAUGCGCGUAGCUUCAACA .(((((((..-...))))))).((((.(((((((..(.((((.(...((((.((((.(.....))))).))))..).)))).)..))))))).))))(((.....)))............ ( -44.60) >consensus UUCAGUUCGCCCUAGAACUGAUAAGGCGACUUUGUGGUUGCGCCUCCGCCAAUUGGGGAAUCUCUCAGGUGGCAUGCCGCACCUGCAGCGUCUCCUUGCGAAAUGCGCGUAGCUUCAACA .(((((((......))))))).((((.(((.(((..(.((((.(...((((.((((((....)))))).))))..).)))).)..))).))).))))(((.....)))............ (-38.82 = -39.10 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:48 2006