
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,068,213 – 12,068,561 |
| Length | 348 |
| Max. P | 0.984000 |

| Location | 12,068,213 – 12,068,326 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -40.66 |
| Consensus MFE | -33.84 |
| Energy contribution | -34.92 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12068213 113 + 27905053 -----CUGGGCUAAAAUUGGUGGCUGGCAACGCCACUCGGCCUCACACACAACUGGCAAUGCCCCUGUAUUGUGAUUGUGUUGUGUGUGC--GCUCUCGCUCUCUCCCAGUCAGUCCCAC -----(((((........((((((.(....))))))).(((..(((((((((((.(((((((....))))))).)....)))))))))).--)))..........))))).......... ( -41.00) >DroSec_CAF1 4833 107 + 1 -----CUGGGCUAAAAUUGGUGGCUGGCAACGCCGCUCGGCCUCACACACAGCUGGCAAUGCCCCUGUAUUGUGAUUGU------UGUGC--GCUCUCGCUCUCUCCCAGUCAGUUCCAC -----(((((........((((((.(....))))))).(((..((((.((((.(.(((((((....))))))).)))))------)))).--)))..........))))).......... ( -32.60) >DroSim_CAF1 5259 113 + 1 -----CUGGACUAAAAUUGGUGGCUGGCAACGCCGCUCGGCCUCACACACAGCUGGCAAUGCCCCUGUAUUGUGAUUGUGUUGUGUGUGC--GCUCUCGCUCUCUCCCAGUCAGUUCCAC -----.((((((...(((((..(..(((...(((....)))(.(((((((((((.(((((((....))))))).)....)))))))))).--).....)))..)..))))).)).)))). ( -39.20) >DroEre_CAF1 6774 118 + 1 CUGUGCUGGGUUGAAAUUGGUGGCUGGCAACGCCGCUCGGCCUCACACACAGCUGGCAAUGCCAUGGUAUUGUGAUUGUGUUGGGUGUGC--UCUCUCGCUCUCUCCCAGUCAGUUCCAC (((.((((((..((....((((((.(....)))))))..((((((.((((((.(.(((((((....))))))).))))))))))).))((--......))))...)))))))))...... ( -43.70) >DroYak_CAF1 6891 120 + 1 CUGUGCUGGGUUGAAAUUGGUGGCUGGCAACACCGCUCGGCCUCACACACAGCUGGCAAUGCCCCAGUAUUGUGAUUGUGUUGUGUGUGCGCGCUCUCGCUCUCUCCCAGUCAGUCCCAC (((.((((((..((....(((((.((....))))))).((((.(((((((((((.(((((((....))))))).)....)))))))))).).))).....))...)))))))))...... ( -46.80) >consensus _____CUGGGCUAAAAUUGGUGGCUGGCAACGCCGCUCGGCCUCACACACAGCUGGCAAUGCCCCUGUAUUGUGAUUGUGUUGUGUGUGC__GCUCUCGCUCUCUCCCAGUCAGUUCCAC ......((((.....(((((..(..(((...(((....)))..(((((((((((.(((((((....))))))).)....)))))))))).........)))..)..)))))....)))). (-33.84 = -34.92 + 1.08)

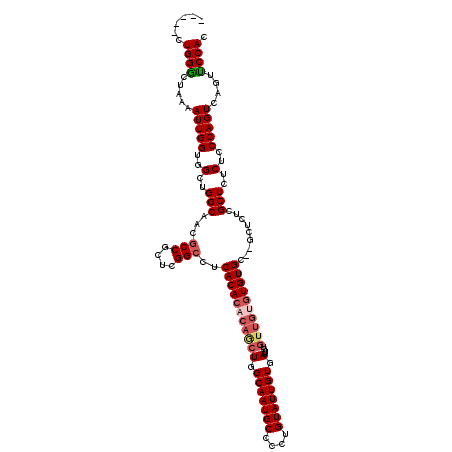

| Location | 12,068,326 – 12,068,441 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -30.50 |
| Energy contribution | -32.26 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12068326 115 - 27905053 AGUGCGAGCGAGAACGAUGCA-AC-GCUGUCUCCCUUCCACGACGCGCACUUAUGUAAAAAGCAUUGAAACUGAACGAGUUUCUCGAUAGCAGCGGGGGUGGGAGAGG-GAG--AGAGCG ..((((..((....)).))))-.(-(((.((((((((((((..(.(((((....)).....(((((((((((.....))))..))))).)).))).).))))..))))-)))--).)))) ( -42.30) >DroSec_CAF1 4940 117 - 1 AGUGCGAGCGAGCACGCUGCAUAC-GCUGUCUCCCUUCCACGACGCGCACUUAUGUAAAAAGCAUUGAAACUGAACGAGUUUCUCGAUAGCAGCGGGGGUGGGAGAGGAGAG--AGAGCG .((((.((((....)))))))).(-(((.((((((((((((..(.(((((....)).....(((((((((((.....))))..))))).)).))).).))))))).))))).--..)))) ( -45.00) >DroSim_CAF1 5372 117 - 1 AGUGCGAGCGAGCACUCUGCAUAC-GCUGUCUCCCUUCCACGACGCGCACUUAUGUAAAAAGCAUUGAAACUGAACGAGUUUCUCGAUAGCAGCGGGGGUGGGAGAGUAGAG--AGAGCG (((((......))))).......(-(((.((((((((((((..(.(((((....)).....(((((((((((.....))))..))))).)).))).).))))))).)..)))--).)))) ( -40.50) >DroEre_CAF1 6892 117 - 1 AGUGCGAGCGAGCACGCUUCAUACAGCUGUCUCCCUUCCGCGACGCGCACUUAUGUAAAAAGCAUUGAAACCGAACGAGUUUCUCGAAAGCAGCGGGGGUGG-AGAGGAGAG--AGAGCG .((((......))))..........(((.((((((((((((..(.(((....((((.....)))).((((((....).))))).........))).).))))-).))).)))--).))). ( -40.60) >DroYak_CAF1 7011 119 - 1 AGUGCGAGCGAGCAAGCUGCAUACAGCUAUCUCCCUCGCGCGACGCGCACUUAUGUAAAAAGCAUUGAAACUGAACGAGUUUCUCGAUAGCAGCGGGGAUGG-UGAGGAGAGAAAGAGCG .((((.(((......)))))))...(((.((((((((((.(..(((((....((((.....)))).((((((.....))))))......)).)))..)...)-))))).))))...))). ( -40.10) >consensus AGUGCGAGCGAGCACGCUGCAUAC_GCUGUCUCCCUUCCACGACGCGCACUUAUGUAAAAAGCAUUGAAACUGAACGAGUUUCUCGAUAGCAGCGGGGGUGGGAGAGGAGAG__AGAGCG .((((......))))..........(((.((((((((((((..(((((....((((.....)))).((((((.....))))))......)).)))...))))))).))))).....))). (-30.50 = -32.26 + 1.76)



| Location | 12,068,403 – 12,068,521 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -52.30 |
| Consensus MFE | -44.48 |
| Energy contribution | -45.60 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12068403 118 + 27905053 UGGAAGGGAGACAGC-GU-UGCAUCGUUCUCGCUCGCACUCACCACGAACGGUGCUCGUACAGGGGUGCAUUUACUUGCGUCUGUGUGCGGGCCUGCCGUUUGGCUGCCUGCUGCCUGCU .((.(((.((.(((.-((-.((...((........))........(((((((((((((((((.((..(((......)))..)).)))))))))..)))))))))).))))))).))).)) ( -49.20) >DroSec_CAF1 5018 119 + 1 UGGAAGGGAGACAGC-GUAUGCAGCGUGCUCGCUCGCACUCACCACGAACGGUGCUCGUACAGGGGUGCAUUUACUUGCGUCUGUGUGCGUGGCUGCCGUUUGGCUGCCUGCUGCCUGCU .....((.((.((((-(...(((((((((......))))......(((((((((((((((((.((..(((......)))..)).)))))).))).))))))))))))).))))).)).)) ( -56.40) >DroSim_CAF1 5450 119 + 1 UGGAAGGGAGACAGC-GUAUGCAGAGUGCUCGCUCGCACUCACCACGAACGGUGCUCGUACAGGGGUGCAUUUACUUGCGUCUGUGUGCGUGCCUGCCGUUUGGCUGCCUGCUGCCUGCU .((.(((.((.(((.-(((.((.((((((......))))))....((((((((((.((((((.((..(((......)))..)).)))))).))..)))))))))))))))))).))).)) ( -57.40) >DroEre_CAF1 6969 119 + 1 CGGAAGGGAGACAGCUGUAUGAAGCGUGCUCGCUCGCACUCAC-UCGCACGGUUCUCGUACAGGGGUGCAUUUACUUGCGUCUGUGUGUGUGCCUACCGUUUGGCUGCCUGCUGCCUGCU (((.((((((...(((......)))...)))...(((((.(((-.((((.(((....((((....))))....)))))))...))).)))))))).)))...(((........))).... ( -39.90) >DroYak_CAF1 7090 120 + 1 CGCGAGGGAGAUAGCUGUAUGCAGCUUGCUCGCUCGCACUCGCCACGAACGGUGCUCGUACAGGGGUGCAUUUACUUGCGUCUGUGUGCGUGCCUGCCGUUUGGCUGCCUGCUGCCUGCU .(((((.(((..(((((....)))))..))).)))))....((((..((((((((.((((((.((..(((......)))..)).)))))).))..)))))))))).....((.....)). ( -58.60) >consensus UGGAAGGGAGACAGC_GUAUGCAGCGUGCUCGCUCGCACUCACCACGAACGGUGCUCGUACAGGGGUGCAUUUACUUGCGUCUGUGUGCGUGCCUGCCGUUUGGCUGCCUGCUGCCUGCU .....((.((.((((.....(((((((((......))))......((((((((((.((((((.((..(((......)))..)).)))))).))..)))))))))))))..)))).)).)) (-44.48 = -45.60 + 1.12)



| Location | 12,068,441 – 12,068,561 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -44.36 |
| Consensus MFE | -41.06 |
| Energy contribution | -41.18 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12068441 120 + 27905053 CACCACGAACGGUGCUCGUACAGGGGUGCAUUUACUUGCGUCUGUGUGCGGGCCUGCCGUUUGGCUGCCUGCUGCCUGCUUCUUGAGCGCCGAACGUUUUAUUUGUGAUUUUAGCACGGC .....(((((((((((((((((.((..(((......)))..)).)))))))))..))))))))...((((((((..(((.....(((((.....))))).....)))....))))).))) ( -47.70) >DroSec_CAF1 5057 120 + 1 CACCACGAACGGUGCUCGUACAGGGGUGCAUUUACUUGCGUCUGUGUGCGUGGCUGCCGUUUGGCUGCCUGCUGCCUGCUUCUUGAGCGCCGAACGUUUUAUUUGUGAUUUUAGCACGGC ..(((((..(((((((((((((.((..(((......)))..)).)))))..(((.(((....))).))).((.....)).....))))))))..))).......(((.......))))). ( -46.80) >DroSim_CAF1 5489 120 + 1 CACCACGAACGGUGCUCGUACAGGGGUGCAUUUACUUGCGUCUGUGUGCGUGCCUGCCGUUUGGCUGCCUGCUGCCUGCUUCUUGAGCGCCGAACGUUUUAUUUGUGAUUUUAGCACGGC .....((((((((((.((((((.((..(((......)))..)).)))))).))..))))))))...((((((((..(((.....(((((.....))))).....)))....))))).))) ( -44.40) >DroEre_CAF1 7009 119 + 1 CAC-UCGCACGGUUCUCGUACAGGGGUGCAUUUACUUGCGUCUGUGUGUGUGCCUACCGUUUGGCUGCCUGCUGCCUGCUUCUUGAGCGCCGAACGUUUUAUUUGUGAUUUUAGCACGGC ...-(((((.(((...((((((.((..(((......)))..)).)))))).)))...((((((((.(((.((.....)).....).)))))))))).......)))))............ ( -35.50) >DroYak_CAF1 7130 120 + 1 CGCCACGAACGGUGCUCGUACAGGGGUGCAUUUACUUGCGUCUGUGUGCGUGCCUGCCGUUUGGCUGCCUGCUGCCUGCUUCUUGAGCGCCGAGCGUUUUAUUUGUGAUUUUAGCACGGU .((((..((((((((.((((((.((..(((......)))..)).)))))).))..)))))))))).((((((((..(((.....((((((...)))))).....)))....))))).))) ( -47.40) >consensus CACCACGAACGGUGCUCGUACAGGGGUGCAUUUACUUGCGUCUGUGUGCGUGCCUGCCGUUUGGCUGCCUGCUGCCUGCUUCUUGAGCGCCGAACGUUUUAUUUGUGAUUUUAGCACGGC .....((((((((((.((((((.((..(((......)))..)).)))))).))..))))))))...((((((((..(((.....(((((.....))))).....)))....))))).))) (-41.06 = -41.18 + 0.12)

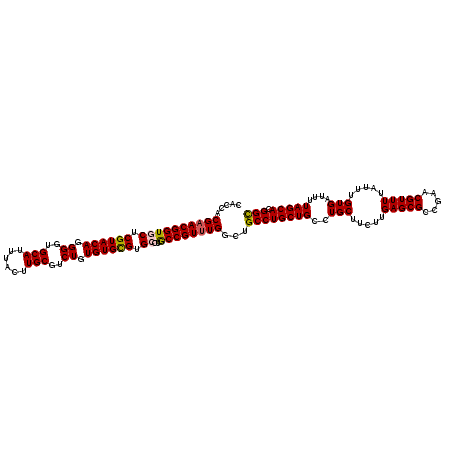

| Location | 12,068,441 – 12,068,561 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -31.36 |
| Energy contribution | -32.00 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12068441 120 - 27905053 GCCGUGCUAAAAUCACAAAUAAAACGUUCGGCGCUCAAGAAGCAGGCAGCAGGCAGCCAAACGGCAGGCCCGCACACAGACGCAAGUAAAUGCACCCCUGUACGAGCACCGUUCGUGGUG ((((((.......))).......(((..(((.((((.....(((((..((.(((.(((....)))..))).))...((.((....))...))....)))))..)))).)))..)))))). ( -40.80) >DroSec_CAF1 5057 120 - 1 GCCGUGCUAAAAUCACAAAUAAAACGUUCGGCGCUCAAGAAGCAGGCAGCAGGCAGCCAAACGGCAGCCACGCACACAGACGCAAGUAAAUGCACCCCUGUACGAGCACCGUUCGUGGUG ((((((.......))).......(((..(((.((((.....(((((.....(((.(((....))).)))..(((.....((....))...)))...)))))..)))).)))..)))))). ( -41.10) >DroSim_CAF1 5489 120 - 1 GCCGUGCUAAAAUCACAAAUAAAACGUUCGGCGCUCAAGAAGCAGGCAGCAGGCAGCCAAACGGCAGGCACGCACACAGACGCAAGUAAAUGCACCCCUGUACGAGCACCGUUCGUGGUG ((((((.......))).......(((..(((.((((........(((........))).....(((((...(((.....((....))...)))...)))))..)))).)))..)))))). ( -37.80) >DroEre_CAF1 7009 119 - 1 GCCGUGCUAAAAUCACAAAUAAAACGUUCGGCGCUCAAGAAGCAGGCAGCAGGCAGCCAAACGGUAGGCACACACACAGACGCAAGUAAAUGCACCCCUGUACGAGAACCGUGCGA-GUG ((((((.......)))........((((.((((((.....)))..((.....)).))).))))...)))...(((.((.((....))...))......((((((.....)))))).-))) ( -30.60) >DroYak_CAF1 7130 120 - 1 ACCGUGCUAAAAUCACAAAUAAAACGCUCGGCGCUCAAGAAGCAGGCAGCAGGCAGCCAAACGGCAGGCACGCACACAGACGCAAGUAAAUGCACCCCUGUACGAGCACCGUUCGUGGCG .(((((.......))).......(((..(((.((((........(((........))).....(((((...(((.....((....))...)))...)))))..)))).)))..))))).. ( -35.20) >consensus GCCGUGCUAAAAUCACAAAUAAAACGUUCGGCGCUCAAGAAGCAGGCAGCAGGCAGCCAAACGGCAGGCACGCACACAGACGCAAGUAAAUGCACCCCUGUACGAGCACCGUUCGUGGUG ((((((.......))).......(((..(((.((((.....(((((..((((.(.(((....))).).)..........((....))...)))...)))))..)))).)))..)))))). (-31.36 = -32.00 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:45 2006