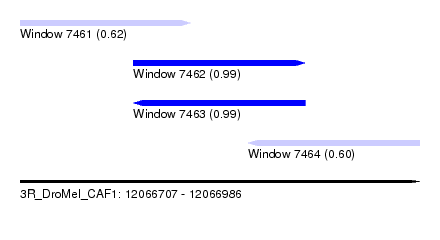
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,066,707 – 12,066,986 |
| Length | 279 |
| Max. P | 0.989642 |

| Location | 12,066,707 – 12,066,826 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.46 |
| Mean single sequence MFE | -25.51 |
| Consensus MFE | -21.70 |
| Energy contribution | -22.70 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12066707 119 + 27905053 UUAUUUCAUUUCAUUUGGCGACUUUUUGGGCCAAAAGAGAUAAGCUGGGCGAAAAGCAGGGAAAAAAA-CGAAAAACAAAAAAUGUUUCAAGGCGAAAUUCGCACAGCAAUCUUGGCAAU .......(((((.((((((..(.....).)))))).)))))..((((.(((((..((...((((....-................))))...))....))))).))))............ ( -27.45) >DroSec_CAF1 3324 117 + 1 UUAUUUCAUUUCAUUUGGCGACUUUUUGGGCCAAAAGAGAUAAGCUGGGCGAAAAGCAGGGAAA-AAA-CGAAAAACAAAAAAUGU-UCAAGGCGAAAUUCGCACAGCAAUCUUGGCAAU .......(((((.((((((..(.....).)))))).)))))..((((.(((((..((...(((.-...-................)-))...))....))))).))))............ ( -26.55) >DroSim_CAF1 3755 118 + 1 UUAUUUCAUUUCAUUUGGCGACUUUUUGGGCCAAAAGAGAUAAGCUGCGCGAAAAGCAGGGAAA-AAA-CGAAAAACAAAAAAUGUUUCAAGGCGAAAUUCGCACAGCAAUCUUGGCAAU .......(((((.((((((..(.....).)))))).)))))..((((.(((((..((...((((-...-................))))...))....))))).))))............ ( -28.31) >DroEre_CAF1 5295 118 + 1 UUAUUUCAUUUAAUUUAGCGACUUUUUGGGCCAAAAGAGAUAAGCUGGGCGAAAAGCAAGGAAA-AAAGCGAAAAACAA-AAAUGUUUCAAGGCGAAAUUCGCACAGCAAUCUUGGCAAU .............................((((....((((..((((.(((((..((...((((-..............-.....))))...))....))))).)))).))))))))... ( -22.91) >DroYak_CAF1 5374 118 + 1 UUUUUUCAUUUCAUUUAGCGACUUUUUGGCCCAAAAGAGAUAAGCUGGGCGAAAAGCAGGGAAA-AAA-CGAAAAACAAAAAAUGUUUCAAGGCGAAAUUCGCACAGCAAUCUUGGCAAU .................((..((((((......))))))....((((.(((((..((...((((-...-................))))...))....))))).)))).......))... ( -22.31) >consensus UUAUUUCAUUUCAUUUGGCGACUUUUUGGGCCAAAAGAGAUAAGCUGGGCGAAAAGCAGGGAAA_AAA_CGAAAAACAAAAAAUGUUUCAAGGCGAAAUUCGCACAGCAAUCUUGGCAAU .......(((((.((((((..........)))))).)))))..((((.(((((..((...((((.....................))))...))....))))).))))............ (-21.70 = -22.70 + 1.00)

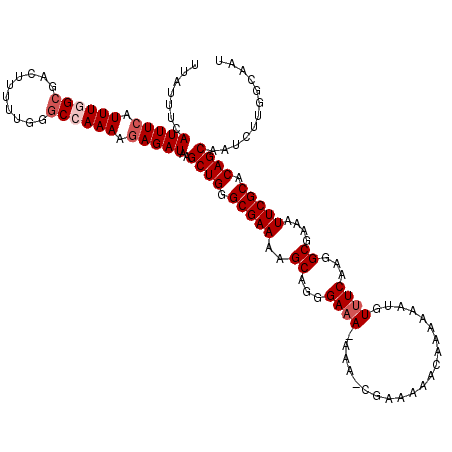
| Location | 12,066,786 – 12,066,906 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -32.40 |
| Energy contribution | -32.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12066786 120 + 27905053 AAAUGUUUCAAGGCGAAAUUCGCACAGCAAUCUUGGCAAUAGCGCAUAAAAUAGCACAGAGCGCACACGAAGCGGGCAAAAAGUACCUGGCAGAUACAGAUACGACUGCCAGUGUGCGGA ...((((((...(((.....)))...((..(((.(((....))((........)).))))..))....))))))........(((((((((((............))))))).))))... ( -32.90) >DroSec_CAF1 3402 119 + 1 AAAUGU-UCAAGGCGAAAUUCGCACAGCAAUCUUGGCAAUAGCGCAUAAAAUAGCACAGAGCGCACACGAAGCGGGCAAAAAGUACCUGGCAGAUACAGAUACGACUGCCAGUGUGCGGA ....((-((...(((.....)))...((.((.(((((......)).))).)).))...))))((((((......((((....(((.(((.......))).)))...)))).))))))... ( -32.90) >DroSim_CAF1 3833 120 + 1 AAAUGUUUCAAGGCGAAAUUCGCACAGCAAUCUUGGCAAUAGCGCAUAAAAUAGCACAGAGCGCACACGAAGCGGGCAAAAAGUACCUGGCAGAUACAGAUACGACUGCCAGUGUGCGGA ...((((((...(((.....)))...((..(((.(((....))((........)).))))..))....))))))........(((((((((((............))))))).))))... ( -32.90) >DroEre_CAF1 5373 120 + 1 AAAUGUUUCAAGGCGAAAUUCGCACAGCAAUCUUGGCAAUAGCGCAUAAAAUAGCACAGAGCGCACACGAAGCGGGCAAAAAGUACCUGGCAGAUACAGAUACGACUGCCAGUGUGCGAA ...((((((...(((.....)))...((..(((.(((....))((........)).))))..))....))))))........(((((((((((............))))))).))))... ( -32.90) >DroYak_CAF1 5452 120 + 1 AAAUGUUUCAAGGCGAAAUUCGCACAGCAAUCUUGGCAAUAGCGCAUAAAAUAGCACAGAGCGCACACGAAGCGGGCAAAAAGUACCUGGCAGAUACAGAUACGACUGCCAGUGUGCGAA ...((((((...(((.....)))...((..(((.(((....))((........)).))))..))....))))))........(((((((((((............))))))).))))... ( -32.90) >consensus AAAUGUUUCAAGGCGAAAUUCGCACAGCAAUCUUGGCAAUAGCGCAUAAAAUAGCACAGAGCGCACACGAAGCGGGCAAAAAGUACCUGGCAGAUACAGAUACGACUGCCAGUGUGCGGA ...((((((...(((.....)))...((..(((.(((....))((........)).))))..))....))))))........(((((((((((............))))))).))))... (-32.40 = -32.60 + 0.20)

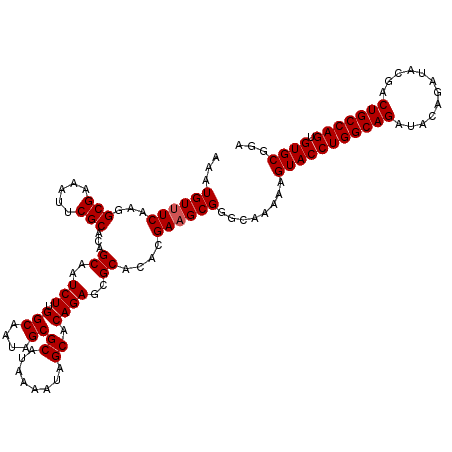

| Location | 12,066,786 – 12,066,906 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -34.22 |
| Energy contribution | -34.22 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12066786 120 - 27905053 UCCGCACACUGGCAGUCGUAUCUGUAUCUGCCAGGUACUUUUUGCCCGCUUCGUGUGCGCUCUGUGCUAUUUUAUGCGCUAUUGCCAAGAUUGCUGUGCGAAUUUCGCCUUGAAACAUUU ..((((((..(((((..(((((((.......)))))))...))))).((.((.((.(((....((((........))))...))))).))..)))))))).................... ( -34.30) >DroSec_CAF1 3402 119 - 1 UCCGCACACUGGCAGUCGUAUCUGUAUCUGCCAGGUACUUUUUGCCCGCUUCGUGUGCGCUCUGUGCUAUUUUAUGCGCUAUUGCCAAGAUUGCUGUGCGAAUUUCGCCUUGA-ACAUUU ..((((((..(((((..(((((((.......)))))))...))))).((.((.((.(((....((((........))))...))))).))..)))))))).............-...... ( -34.30) >DroSim_CAF1 3833 120 - 1 UCCGCACACUGGCAGUCGUAUCUGUAUCUGCCAGGUACUUUUUGCCCGCUUCGUGUGCGCUCUGUGCUAUUUUAUGCGCUAUUGCCAAGAUUGCUGUGCGAAUUUCGCCUUGAAACAUUU ..((((((..(((((..(((((((.......)))))))...))))).((.((.((.(((....((((........))))...))))).))..)))))))).................... ( -34.30) >DroEre_CAF1 5373 120 - 1 UUCGCACACUGGCAGUCGUAUCUGUAUCUGCCAGGUACUUUUUGCCCGCUUCGUGUGCGCUCUGUGCUAUUUUAUGCGCUAUUGCCAAGAUUGCUGUGCGAAUUUCGCCUUGAAACAUUU ((((((((..(((((..(((((((.......)))))))...))))).((.((.((.(((....((((........))))...))))).))..)))))))))).................. ( -36.30) >DroYak_CAF1 5452 120 - 1 UUCGCACACUGGCAGUCGUAUCUGUAUCUGCCAGGUACUUUUUGCCCGCUUCGUGUGCGCUCUGUGCUAUUUUAUGCGCUAUUGCCAAGAUUGCUGUGCGAAUUUCGCCUUGAAACAUUU ((((((((..(((((..(((((((.......)))))))...))))).((.((.((.(((....((((........))))...))))).))..)))))))))).................. ( -36.30) >consensus UCCGCACACUGGCAGUCGUAUCUGUAUCUGCCAGGUACUUUUUGCCCGCUUCGUGUGCGCUCUGUGCUAUUUUAUGCGCUAUUGCCAAGAUUGCUGUGCGAAUUUCGCCUUGAAACAUUU ..((((((..(((((..(((((((.......)))))))...))))).((.((.((.(((....((((........))))...))))).))..)))))))).................... (-34.22 = -34.22 + -0.00)

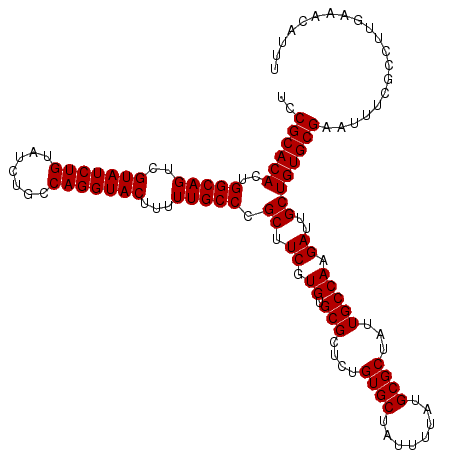

| Location | 12,066,866 – 12,066,986 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -30.50 |
| Energy contribution | -30.66 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12066866 120 - 27905053 ACCCCUCUUUGUUAAAACAUUUUUGUUUAUUUCAGCGGCGCGGUCGUAUCUGUAUCUCAGAUGUUGGCUGACUGGUUCGCUCCGCACACUGGCAGUCGUAUCUGUAUCUGCCAGGUACUU ..............(((((....)))))......((((.((((...((((((.....))))))...(((....))))))).))))((.(((((((............))))))))).... ( -32.80) >DroSec_CAF1 3481 120 - 1 ACCCCUCUUUGUUAAAACAUUUUUGUUUAUUUCAGCGGCGCGGUCGUAUCUGUAUCUCAGAUGUUGGCUGACUGGUUCGCUCCGCACACUGGCAGUCGUAUCUGUAUCUGCCAGGUACUU ..............(((((....)))))......((((.((((...((((((.....))))))...(((....))))))).))))((.(((((((............))))))))).... ( -32.80) >DroSim_CAF1 3913 118 - 1 ACCCCUCUUUGUUA-AACAUUUUUGU-UAUUUCAGCGGCGCGGUCGUAUCUGUAUCUCAGAUGUUGGCUGACUGGUUCGCUCCGCACACUGGCAGUCGUAUCUGUAUCUGCCAGGUACUU ..............-((((....)))-)......((((.((((...((((((.....))))))...(((....))))))).))))((.(((((((............))))))))).... ( -31.90) >DroEre_CAF1 5453 120 - 1 CCCCCUCUUUGAGGAAACAUUUUUGUUUAUUUCAGCGGCGCGCUCGUAUCUGUAUCUCAGAUGUUGGCUGACUGGUUCGCUUCGCACACUGGCAGUCGUAUCUGUAUCUGCCAGGUACUU ...((((...))))(((((....)))))......((((.(((((..((((((.....))))))..))).((.....)))).))))((.(((((((............))))))))).... ( -36.00) >DroYak_CAF1 5532 120 - 1 CCCCCUCUUUGUGGAAACAUUUUUGUUUAUUUCAGCGGCGCGCUCGUAUCUGUAUCUCAGAUGUUGGCUGACUGGUUCGCUUCGCACACUGGCAGUCGUAUCUGUAUCUGCCAGGUACUU ..........(((....)))..............((((.(((((..((((((.....))))))..))).((.....)))).))))((.(((((((............))))))))).... ( -33.40) >consensus ACCCCUCUUUGUUAAAACAUUUUUGUUUAUUUCAGCGGCGCGGUCGUAUCUGUAUCUCAGAUGUUGGCUGACUGGUUCGCUCCGCACACUGGCAGUCGUAUCUGUAUCUGCCAGGUACUU ..............(((((....)))))......((((.(((....((((((.....))))))..((((....))))))).))))((.(((((((............))))))))).... (-30.50 = -30.66 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:37 2006