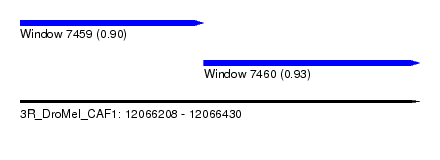
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,066,208 – 12,066,430 |
| Length | 222 |
| Max. P | 0.932393 |

| Location | 12,066,208 – 12,066,310 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -20.06 |
| Energy contribution | -21.06 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12066208 102 + 27905053 CCAUUA------------------UAACAGUAGGGUUUCCUUCAACCAUCUGGUACAGUCAUCCCUUCCCCGCGAGAUACAUAGUACCCGGGGACCCAUUAUACCGCCGUUGAUAACCAA ......------------------.........((((....(((((....(((((.(((.......((((((.(.(.(((...)))))))))))...))).)))))..))))).)))).. ( -21.20) >DroSec_CAF1 2807 120 + 1 CCAUUAUAUUAUCCAAAUAUUCCAUAACAGUACGGUUGCCUUCAACCAUCUGGUACAGUCAUCCCUUCCCCGCGAGAUACAUAGUACCCGGGGACCCAUUGUACCGCCGUUGAUAACCAA .................................(((((...(((((....(((((((((.......((((((.(.(.(((...)))))))))))...)))))))))..)))))))))).. ( -27.60) >DroSim_CAF1 3238 120 + 1 CCAUUAUAUUAUCCAACUAUUCCAUAACAGUAAGGUUUCCUUCAACCAUCUGGUACAGUCAUCCCUUCCCCGCGAGAUACAUAGUACCCGGGGACCCAUUGUACCGCCGUUGAUAACCAA .................................((((....(((((....(((((((((.......((((((.(.(.(((...)))))))))))...)))))))))..))))).)))).. ( -27.40) >DroEre_CAF1 4776 119 + 1 CA-UUAUAUUGUCCAACUACUCCAUAACAGUAAGGCUUCCCUCUACCAUCUGGUACAGUCGUCCCUUCCCCACGAUAUACAUGGUACCCGGGGACUCAUUGUGCCACAGUUGAUAACCAA ..-.....(((((.((((...........((((((....))).)))....(((((((((.((((((...(((.(.....).))).....))))))..))))))))).))))))))).... ( -29.20) >DroYak_CAF1 4854 120 + 1 CCUUUAGAUUGUCCAACUACUCCAUAACAGUAAGGCUUCUCUCUACCAUCUGGUACAGUCAUCCCUUCCCCGCGAGAUACAUAGUACCCGUGGACCCAUUGUACCACAGUUGAUAACCAA ......(.(((((.((((........(((((((((.....((.((((....)))).)).....))))..(((((.(.(((...))).))))))....))))).....))))))))).).. ( -21.42) >consensus CCAUUAUAUUAUCCAACUACUCCAUAACAGUAAGGUUUCCUUCAACCAUCUGGUACAGUCAUCCCUUCCCCGCGAGAUACAUAGUACCCGGGGACCCAUUGUACCGCCGUUGAUAACCAA .................................((((....(((((....(((((((((.......((((((.(..((.....))..)))))))...)))))))))..))))).)))).. (-20.06 = -21.06 + 1.00)



| Location | 12,066,310 – 12,066,430 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -33.39 |
| Consensus MFE | -31.82 |
| Energy contribution | -32.02 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12066310 120 + 27905053 UAGCGAUAGCCUCGACCAGUUGCAUGUCCGUCAUCAGCUGGACGAGAACUCGAACUACCUUUGUACCCCUGUCGAUGGGGAAUAAAAAAUAAACCCCAAUCUCGUUCGCCUCGAAGUAAU ..((((.........(((((((.(((.....))))))))))((((((..((((..(((....)))......))))(((((.............))))).))))))))))........... ( -32.82) >DroSec_CAF1 2927 120 + 1 UAGCGAUAGCCUCGACCAGUUGCAUGUCCGUCAUCAGCUGGACGAGAACCCGAACUACCUUUGUACCCCUGUCGAUGGGGAAUAAAAAAUAAACCCCAAUCUCGUUCGCCUCGAAGUAAU ..((((....((((.(((((((.(((.....)))))))))).)))).............(((((.((((.......)))).)))))...................))))........... ( -32.30) >DroSim_CAF1 3358 120 + 1 UAGCGAUAGCCUCGACCAGUUGCAUGUCCGUCAUCAGCUGGACGAGAACCCGAACUACCUUUGUACCCCUGUCGAUGGGGAAUAAAAAAUAAACCCCAAUCUCGUUCGCCUCGAAGUAAU ..((((....((((.(((((((.(((.....)))))))))).)))).............(((((.((((.......)))).)))))...................))))........... ( -32.30) >DroEre_CAF1 4895 120 + 1 UAGCGAUAGCCUCGACCAGUUGCAUGUCGGUCAUCAGCUGGACGAGAACCCGAACUACCUUUGUACCCCUGUCGAUGGGGAAUAAAAAAUAAACCCCAAUCUCGUUCGCCUCGAAGUAAU ..((((....((((.(((((((.(((.....)))))))))).)))).............(((((.((((.......)))).)))))...................))))........... ( -32.30) >DroYak_CAF1 4974 120 + 1 UAGCGAUGGCCUCGACCAGUUGCAUGUCCGUCAUCAGCUGGACGAGAACCCGAACUACCUUUGGACCCCUGUCGAUGGGGAAUAAAAAAUAAACCCCAAUCUCGUUCGCCUCAAAGUAAU ....((.(((((((.(((((((.(((.....)))))))))).)))).....((((........(((....)))(((((((.............)))).)))..)))))))))........ ( -37.22) >consensus UAGCGAUAGCCUCGACCAGUUGCAUGUCCGUCAUCAGCUGGACGAGAACCCGAACUACCUUUGUACCCCUGUCGAUGGGGAAUAAAAAAUAAACCCCAAUCUCGUUCGCCUCGAAGUAAU ..((((....((((.(((((((.(((.....)))))))))).)))).............(((((.((((.......)))).)))))...................))))........... (-31.82 = -32.02 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:33 2006