
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,065,477 – 12,065,716 |
| Length | 239 |
| Max. P | 0.999961 |

| Location | 12,065,477 – 12,065,597 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -34.60 |
| Energy contribution | -34.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12065477 120 - 27905053 GCGACUGUGACGUUGAUGGUGCUGUUACAAUGCCGCUGGCGGCGCUGUUGGCGAUUUGAAUAUGUUAUGAUGUUGAUGUUGCAUGUUGGCCCGCGUCGCCAACUGGUUUUAUUUUUAUUC (((((.....(((..(((((((((((((......).))))))))))))..)))...(.(((((......))))).).)))))..((((((.......))))))................. ( -37.00) >DroSec_CAF1 2066 120 - 1 GCGACAGUGACGUUGAUGGUGCUGUUACAAUGCCGCUGGCGGCGCUGUUGGCGAUUUGAAUAUGUUAUGAUGUUGAUGUUGCAUGUUGGCCCGCGUCGCCAACUGGUUUUAUUUUUAUUC ((((((....(((..(((((((((((((......).))))))))))))..)))...(.(((((......))))).)))))))..((((((.......))))))................. ( -38.50) >DroSim_CAF1 2535 120 - 1 GCGACAGUGACGUUGAUGGUGCUGUUACAAUGCCGCUGGCGGCGCUGUUGGCGAUUUGAAUAUGUUAUGAUGUUGAUGUUGCAUGUUGGCCCGCGUCGCCAACUGGUUUUAUUUUUAUUC ((((((....(((..(((((((((((((......).))))))))))))..)))...(.(((((......))))).)))))))..((((((.......))))))................. ( -38.50) >DroEre_CAF1 4043 120 - 1 GUGACUGUGACGUUGAUGUUGCUGUUACAAUGCUGCUGGCGGCGCUGUUGGCGAUUUGAAUAUGUUAUGAUGUUGAUGUUGCAUGUUGGCCCGCGUAGCCAACUGGUUUUGUUUUUAUUC (((((.(..(((....)))..).)))))...(((((..((((.((((...(((((.(.(((((......))))).).)))))....)))))))))))))..................... ( -35.30) >DroYak_CAF1 4079 120 - 1 GUGACUGUGACGUUGAUGCCGCUGUUACAAUGCCGCUGGCGGCGCUGUUGGCGAUUUGAAUAUGUUAUGAUGUUGAUGUUGCAUGUUGGCCCGCGUCGCCAACUGGUUUUAUUUUUAUUC (((((.(((.((....)).))).)))))...((((.(((((((((.((..(((...((...((((((......))))))..)))))..))..)))))))))..))))............. ( -36.20) >consensus GCGACUGUGACGUUGAUGGUGCUGUUACAAUGCCGCUGGCGGCGCUGUUGGCGAUUUGAAUAUGUUAUGAUGUUGAUGUUGCAUGUUGGCCCGCGUCGCCAACUGGUUUUAUUUUUAUUC (((((.....(((..(((((((((((((......).))))))))))))..)))...(.(((((......))))).).)))))..((((((.......))))))................. (-34.60 = -34.60 + 0.00)

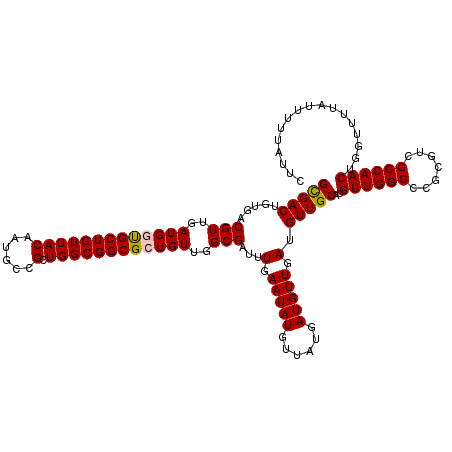

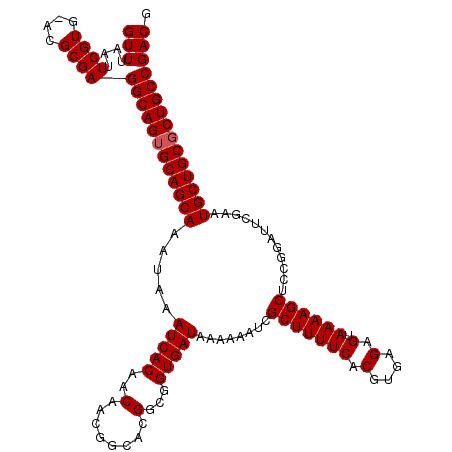
| Location | 12,065,517 – 12,065,637 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -24.62 |
| Energy contribution | -26.06 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12065517 120 + 27905053 AACAUCAACAUCAUAACAUAUUCAAAUCGCCAACAGCGCCGCCAGCGGCAUUGUAACAGCACCAUCAACGUCACAGUCGCCGUCGGCAGCGCAGCAUUCGAAUCCGGAGCUUUUACUCUC ...................................((((.(((.((((((((((.((............)).))))).))))).))).))))(((.((((....)))))))......... ( -32.60) >DroSec_CAF1 2106 120 + 1 AACAUCAACAUCAUAACAUAUUCAAAUCGCCAACAGCGCCGCCAGCGGCAUUGUAACAGCACCAUCAACGUCACUGUCGCCGUCGGCAGCGCAGCAUUCGAAUCCGGAGCUUUUACUCUC ...................................((((.(((.(((((((.((.((............)).)).)).))))).))).))))(((.((((....)))))))......... ( -28.80) >DroSim_CAF1 2575 120 + 1 AACAUCAACAUCAUAACAUAUUCAAAUCGCCAACAGCGCCGCCAGCGGCAUUGUAACAGCACCAUCAACGUCACUGUCGCCGUCGGCAGGGCAGCAUUCGAAUCCGGAGCUUUUACUCUC ............................(((.((((.((((....)))).))))...................((((((....)))))))))(((.((((....)))))))......... ( -28.10) >DroEre_CAF1 4083 120 + 1 AACAUCAACAUCAUAACAUAUUCAAAUCGCCAACAGCGCCGCCAGCAGCAUUGUAACAGCAACAUCAACGUCACAGUCACCGUCGGCAGCGCAGCAUUCGGAACCGGAGCUUUUACUCUC ...................................((((.(((.((.(.(((((.((............)).)))))..).)).))).))))(((.((((....)))))))......... ( -23.20) >DroYak_CAF1 4119 119 + 1 AACAUCAACAUCAUAACAUAUUCAAAUCGCCAACAGCGCCGCCAGCGGCAUUGUAACAGCGGCAUCAACGUCACAGUCACCGUCGGCAGCGCAGCAUUG-GGACCGGGGCUUUUACUCUC ............................(((....((((.(((.((((.(((((....(((.......))).)))))..)))).))).))))....(((-....)))))).......... ( -29.60) >consensus AACAUCAACAUCAUAACAUAUUCAAAUCGCCAACAGCGCCGCCAGCGGCAUUGUAACAGCACCAUCAACGUCACAGUCGCCGUCGGCAGCGCAGCAUUCGAAUCCGGAGCUUUUACUCUC ...................................((((.(((.((((((((((.((............)).))))).))))).))).))))(((.((((....)))))))......... (-24.62 = -26.06 + 1.44)

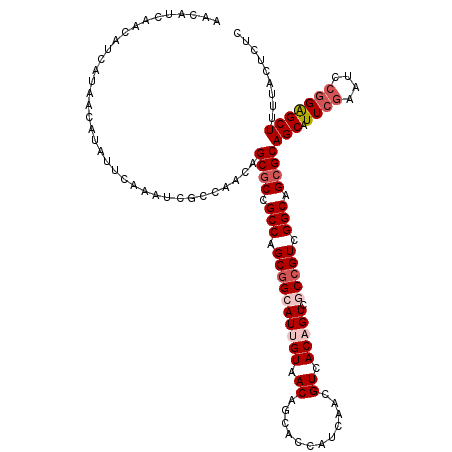

| Location | 12,065,597 – 12,065,716 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.82 |
| Mean single sequence MFE | -41.54 |
| Consensus MFE | -40.34 |
| Energy contribution | -40.54 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.91 |
| SVM RNA-class probability | 0.999961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12065597 119 - 27905053 GUUUAAUUCGUG-ACGCGAGGCAGUGCAGCAAAUAAAUCACAACAACGGCACGGCGGUGAUAAAAAAUCGCUUUUCACGUGAGAGUAAAAGCUCCGGAUUCGAAUGCUGCGCUGCCGACG (((....(((..-...)))((((((((((((....((((......(((....((((((........)))))).....)))(.((((....))))).))))....))))))))))))))). ( -42.30) >DroSec_CAF1 2186 119 - 1 GUUUAAUUCGUG-ACGCGAGGCAGUGCAGCAAAUAAAUCACAACAACGGCACGGCGGUGAUAAAAAAUCGCUUUUCACGUGAGAGUAAAAGCUCCGGAUUCGAAUGCUGCGCUGCCGACG (((....(((..-...)))((((((((((((....((((......(((....((((((........)))))).....)))(.((((....))))).))))....))))))))))))))). ( -42.30) >DroSim_CAF1 2655 119 - 1 GUUUAAUUCGUG-ACGCGAGGCAGUGCAGCAAAUAAAUCACAACAACGGCACGGCGGUGAUAAAAAAUCGCUUUUCACGUGAGAGUAAAAGCUCCGGAUUCGAAUGCUGCCCUGCCGACG (((....(((..-...)))(((((.((((((....((((......(((....((((((........)))))).....)))(.((((....))))).))))....)))))).)))))))). ( -37.90) >DroEre_CAF1 4163 120 - 1 GUUUAAUUCGUGGACGCGAGGCAGUGCAGCAAAUAAAUCACAACAACGGCACGGCGGUGAUAAAAAAUCGCUUUUCACGUGAGAGUAAAAGCUCCGGUUCCGAAUGCUGCGCUGCCGACG (((....((((....))))((((((((((((....((((......(((....((((((........)))))).....)))(.((((....))))))))).....))))))))))))))). ( -42.90) >DroYak_CAF1 4199 119 - 1 GUUAUACUCGUGGACGCGAGGCAGUGCAGCAAAUAAAUCACAACAACGGCACGGCGGUGAUAAAAAAUCGCUUUUCACGUGAGAGUAAAAGCCCCGGUCC-CAAUGCUGCGCUGCCGACG (((....((((....))))((((((((((((................(((..((((((........)))))).....(....).......)))..(....-)..))))))))))))))). ( -42.30) >consensus GUUUAAUUCGUG_ACGCGAGGCAGUGCAGCAAAUAAAUCACAACAACGGCACGGCGGUGAUAAAAAAUCGCUUUUCACGUGAGAGUAAAAGCUCCGGAUUCGAAUGCUGCGCUGCCGACG (((....((((....))))((((((((((((.....(((((..(........)...)))))........(((((((.(....).).))))))............))))))))))))))). (-40.34 = -40.54 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:31 2006