
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,064,888 – 12,065,087 |
| Length | 199 |
| Max. P | 0.972466 |

| Location | 12,064,888 – 12,065,008 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.66 |
| Mean single sequence MFE | -43.20 |
| Consensus MFE | -38.60 |
| Energy contribution | -38.56 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12064888 120 + 27905053 CAAGACUAUGCAAAGCGGACACCUGUUUUUGCAACACGAUCGCUAUUGUUGUAGCGGGUCUGCCGAUGUUGCUGCCGUGGAUGUUGCAGCAGUAGCAGUAUAAUGGUGGUGGUGCUGCUU ...........(((((((....))))))).(((.(((.(((((((((((.....(((.....))).(((((((((.(..(...)..).)))))))))..))))))))))).))).))).. ( -41.60) >DroSec_CAF1 1466 120 + 1 CAAGACUAUGCAAAGCGGACACCUGUUUUUGCAACACGAUCGCUAUUGUUGUAGCGGGGCUGCCGAUGUUGCUGCCGUGGAUGUUGCAGCAGUAGCAGUAUAAUGGUGGUGGUGCUGCUG ...(((..((((((((((....))).))))))).(((..(((((((....)))))))(((.((.......)).))))))...))).((((((((.((.(((....))).)).)))))))) ( -43.40) >DroSim_CAF1 1935 120 + 1 CAAGACUAUGCAAAGCGGACACCUGUUUUUGCAACACGAUCGCUAUUGUUGUAGCGGGGCUGCCGAUGUUGCUGCCGUGGAUGUUGCAGCAGUAGCAGUAUAAUGGUGGUGGUGCUGCUG ...(((..((((((((((....))).))))))).(((..(((((((....)))))))(((.((.......)).))))))...))).((((((((.((.(((....))).)).)))))))) ( -43.40) >DroEre_CAF1 3450 119 + 1 AAAGACUAUACGAAGCGGACACCUGUUUUUGCAACACGAUCGCUAUUGUUGUAGCGGGGCUGU-GGUGUUGCUGCCGUGGAAGUUUCAGCAGUAGCAGUACAAUGAUGGUGGUGCUGCUG ..(((((.((((.((((.((((((((....)))(((...(((((((....)))))))...)))-)))))))))..))))..)))))...((((((((.(((.......))).)))))))) ( -43.20) >DroYak_CAF1 3500 119 + 1 CAAGACUUUGCGAAGCGGACACCUGUUUUUGCAACACGAUCGCUAUUGUUGUUGAGCGGCUGC-GGUGUUGCUGCCGUGGAUGGUGCAGCAGUAGCAGUACAAUGGUGGUGGUGCUGCUG ...........(((((((....))))))).(((.(((.(((((((((((((.....))(((((-..((((((.((((....))))))))))...)))))))))))))))).))).))).. ( -44.40) >consensus CAAGACUAUGCAAAGCGGACACCUGUUUUUGCAACACGAUCGCUAUUGUUGUAGCGGGGCUGCCGAUGUUGCUGCCGUGGAUGUUGCAGCAGUAGCAGUAUAAUGGUGGUGGUGCUGCUG ...........(((((((....))))))).(((.(((.(((((((((((.((((.....))))...(((((((((.((.......)).)))))))))..))))))))))).))).))).. (-38.60 = -38.56 + -0.04)



| Location | 12,064,888 – 12,065,008 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.66 |
| Mean single sequence MFE | -30.66 |
| Consensus MFE | -25.32 |
| Energy contribution | -26.52 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12064888 120 - 27905053 AAGCAGCACCACCACCAUUAUACUGCUACUGCUGCAACAUCCACGGCAGCAACAUCGGCAGACCCGCUACAACAAUAGCGAUCGUGUUGCAAAAACAGGUGUCCGCUUUGCAUAGUCUUG ..(((((((.............(((((..((((((..........)))))).....)))))...(((((......)))))...))))))).....((((.....((...)).....)))) ( -32.00) >DroSec_CAF1 1466 120 - 1 CAGCAGCACCACCACCAUUAUACUGCUACUGCUGCAACAUCCACGGCAGCAACAUCGGCAGCCCCGCUACAACAAUAGCGAUCGUGUUGCAAAAACAGGUGUCCGCUUUGCAUAGUCUUG .(((.(((((..............((....))((((((((....(((.((.......)).))).(((((......)))))...))))))))......)))))..)))............. ( -32.10) >DroSim_CAF1 1935 120 - 1 CAGCAGCACCACCACCAUUAUACUGCUACUGCUGCAACAUCCACGGCAGCAACAUCGGCAGCCCCGCUACAACAAUAGCGAUCGUGUUGCAAAAACAGGUGUCCGCUUUGCAUAGUCUUG .(((.(((((..............((....))((((((((....(((.((.......)).))).(((((......)))))...))))))))......)))))..)))............. ( -32.10) >DroEre_CAF1 3450 119 - 1 CAGCAGCACCACCAUCAUUGUACUGCUACUGCUGAAACUUCCACGGCAGCAACACC-ACAGCCCCGCUACAACAAUAGCGAUCGUGUUGCAAAAACAGGUGUCCGCUUCGUAUAGUCUUU ((((((...((.((....))...))...))))))..(((...((((.(((.(((((-.......(((((......)))))....((((.....)))))))))..)))))))..))).... ( -28.90) >DroYak_CAF1 3500 119 - 1 CAGCAGCACCACCACCAUUGUACUGCUACUGCUGCACCAUCCACGGCAGCAACACC-GCAGCCGCUCAACAACAAUAGCGAUCGUGUUGCAAAAACAGGUGUCCGCUUCGCAAAGUCUUG .(((.(((((.........(((....)))(((.((((.(((...(((.((......-)).)))(((..........)))))).)))).)))......)))))..)))............. ( -28.20) >consensus CAGCAGCACCACCACCAUUAUACUGCUACUGCUGCAACAUCCACGGCAGCAACAUCGGCAGCCCCGCUACAACAAUAGCGAUCGUGUUGCAAAAACAGGUGUCCGCUUUGCAUAGUCUUG ..(((((((.............(((((..((((((..........)))))).....)))))...(((((......)))))...)))))))......((((....))))............ (-25.32 = -26.52 + 1.20)

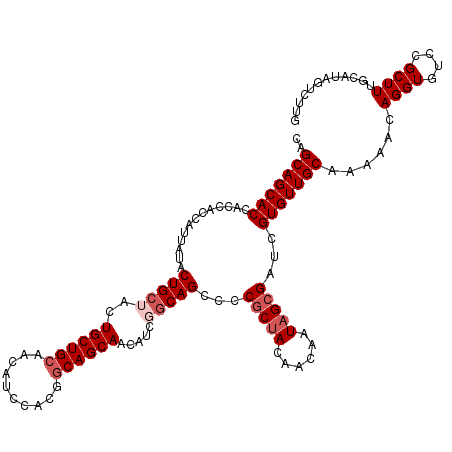

| Location | 12,064,928 – 12,065,047 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -45.91 |
| Consensus MFE | -36.18 |
| Energy contribution | -36.78 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12064928 119 + 27905053 CGCUAUUGUUGUAGCGGGUCUGCCGAUGUUGCUGCCGUGGAUGUUGCAGCAGUAGCAGUAUAAUGGUGGUGGUGCUGCUUCUGAAAAGUCCUCCUACCCAACCACC-UCUGCUUUCAUCU .((((((((((((((..(((..(.(.........).)..)))))))))))))))))((((....((((((((((......((....))......))))..))))))-..))))....... ( -39.60) >DroSec_CAF1 1506 120 + 1 CGCUAUUGUUGUAGCGGGGCUGCCGAUGUUGCUGCCGUGGAUGUUGCAGCAGUAGCAGUAUAAUGGUGGUGGUGCUGCUGCUGAAAAGUCCUCCUCCCCAACCCCCUUCUGCUUUCAUCU .(((((....)))))(((((.((.......)).)))(.((((....(((((((((((.(((.......))).)))))))))))....)))).)....))..................... ( -42.10) >DroSim_CAF1 1975 120 + 1 CGCUAUUGUUGUAGCGGGGCUGCCGAUGUUGCUGCCGUGGAUGUUGCAGCAGUAGCAGUAUAAUGGUGGUGGUGCUGCUGCUGAAAAGUCCUCCUCCCCAACCCCCUUCUGCUUUCAUCU .(((((....)))))(((((.((.......)).)))(.((((....(((((((((((.(((.......))).)))))))))))....)))).)....))..................... ( -42.10) >DroEre_CAF1 3490 119 + 1 CGCUAUUGUUGUAGCGGGGCUGU-GGUGUUGCUGCCGUGGAAGUUUCAGCAGUAGCAGUACAAUGAUGGUGGUGCUGCUGCUGAAAAGUCCUCUUCCCCAACACCCCGCUCCUUUCAUCU .(((((....)))))(((((.(.-(((((((..(..(.(((..((((((((((((((.(((.......))).))))))))))))))..))).)..)..)))))))).)))))........ ( -58.60) >DroYak_CAF1 3540 119 + 1 CGCUAUUGUUGUUGAGCGGCUGC-GGUGUUGCUGCCGUGGAUGGUGCAGCAGUAGCAGUACAAUGGUGGUGGUGCUGCUGCUGAAAAGUCCUCCUCCCCGACACCCCUCUCCACUCAUCU ............((((.((..(.-(((((((.......((((....(((((((((((.(((.......))).)))))))))))....)))).......))))))).)...)).))))... ( -47.14) >consensus CGCUAUUGUUGUAGCGGGGCUGCCGAUGUUGCUGCCGUGGAUGUUGCAGCAGUAGCAGUAUAAUGGUGGUGGUGCUGCUGCUGAAAAGUCCUCCUCCCCAACCCCCCUCUGCUUUCAUCU ((((((....)))))).(((.((.......)).)))(.((((....(((((((((((.(((.......))).)))))))))))....)))).)........................... (-36.18 = -36.78 + 0.60)



| Location | 12,064,968 – 12,065,087 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -34.69 |
| Consensus MFE | -27.82 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.941025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12064968 119 + 27905053 AUGUUGCAGCAGUAGCAGUAUAAUGGUGGUGGUGCUGCUUCUGAAAAGUCCUCCUACCCAACCACC-UCUGCUUUCAUCUGCUGCUUCUGCAAGUCGGACCAUUUAUUCAGCAUUCAAAA ((((((((((((.(((((......((((((((((......((....))......))))..))))))-.))))).....))))))).((((.....))))..........)))))...... ( -35.30) >DroSec_CAF1 1546 120 + 1 AUGUUGCAGCAGUAGCAGUAUAAUGGUGGUGGUGCUGCUGCUGAAAAGUCCUCCUCCCCAACCCCCUUCUGCUUUCAUCUGCUGCUUCUGCAACUCGGGCCAUUUAUUCAGCAUUCAAAA (((((((((((((((((.(((.......))).)))))))))))....((((...................((........))(((....)))....))))........))))))...... ( -34.10) >DroSim_CAF1 2015 120 + 1 AUGUUGCAGCAGUAGCAGUAUAAUGGUGGUGGUGCUGCUGCUGAAAAGUCCUCCUCCCCAACCCCCUUCUGCUUUCAUCUGCUGCUUCUGCAACUCGGGCCAUUUAUUCAGCAUUCAAAA (((((((((((((((((.(((.......))).)))))))))))....((((...................((........))(((....)))....))))........))))))...... ( -34.10) >DroEre_CAF1 3529 120 + 1 AAGUUUCAGCAGUAGCAGUACAAUGAUGGUGGUGCUGCUGCUGAAAAGUCCUCUUCCCCAACACCCCGCUCCUUUCAUCUGCUGCUUCUGCCACUCGGGCCAUUUAUUCAGCAUUCAAAA ...((((((((((((((.(((.......))).)))))))))))))).................................(((((.....(((.....)))........)))))....... ( -34.82) >DroYak_CAF1 3579 120 + 1 AUGGUGCAGCAGUAGCAGUACAAUGGUGGUGGUGCUGCUGCUGAAAAGUCCUCCUCCCCGACACCCCUCUCCACUCAUCUGCUGCUUCUGCCACUCGGGCCAUUUAUUCAGCAUUCAAAA (((((((((((((((((.(((.......))).)))))))))))....(((.........))).........))).))).(((((.....(((.....)))........)))))....... ( -35.12) >consensus AUGUUGCAGCAGUAGCAGUAUAAUGGUGGUGGUGCUGCUGCUGAAAAGUCCUCCUCCCCAACCCCCCUCUGCUUUCAUCUGCUGCUUCUGCAACUCGGGCCAUUUAUUCAGCAUUCAAAA (((((((((((((((((.(((.......))).)))))))))))....((((...................((........)).((....)).....))))........))))))...... (-27.82 = -28.62 + 0.80)

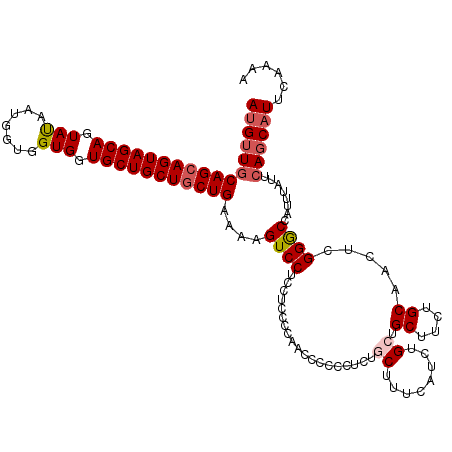
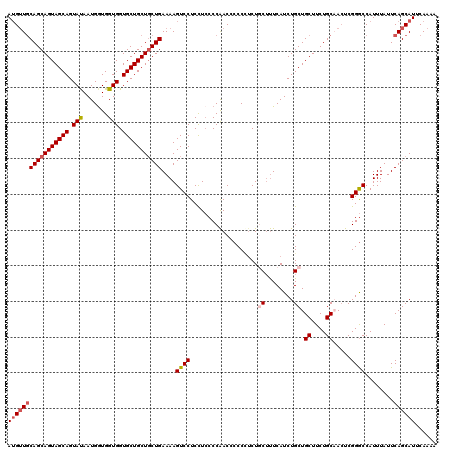
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:25 2006