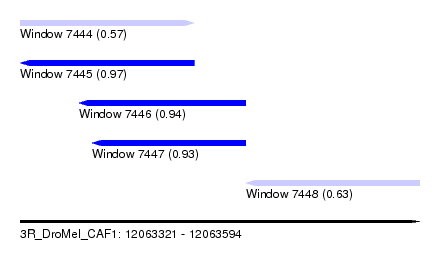
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,063,321 – 12,063,594 |
| Length | 273 |
| Max. P | 0.968892 |

| Location | 12,063,321 – 12,063,440 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -37.13 |
| Consensus MFE | -35.79 |
| Energy contribution | -35.79 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12063321 119 + 27905053 GUAGUUUUUGCAAGAGGAAGAGAGCAGGAAGUGAAUGAGAAUUCUCUUUCCUUUUGUUUUGCGGCGAAGCGAUCCGCA-AGCAAUACGUCGAAUUCGCCGCGAACGGAUCGCUUUUGUUU ((.((....(((((((((((((((.................))))).))))))))))...)).))(((((((((((..-.((....((.......))..))...)))))))))))..... ( -36.93) >DroSec_CAF1 35353 120 + 1 GUAGUUUUUGCAAGAGGAAGAGAGCAGGAAGUGAAUGAGAAUUCUCUUUCCUUUUGUUUUGCGGCGAAGCGAUCCGCAAAGCUAUAUGUCGAAUUCGCCGCGAACGGAUCGCUUUUGUUU ((.((....(((((((((((((((.................))))).))))))))))...)).))(((((((((((....((.....(.((....)).)))...)))))))))))..... ( -35.73) >DroSim_CAF1 44835 120 + 1 GUAGUUUUUGCAAGAGGAAGAGAGCAGGAAGUGAAUGAGAAUUCUCUUUCCUUUUGUUUUGCGGCGAAGCGAUCCGCAAAGCGAUAUGUCGAAUUCGCCGCGAACGGAUCGCUUUUGUUU ((.((....(((((((((((((((.................))))).))))))))))...)).))(((((((((((....((((..........))))......)))))))))))..... ( -38.73) >consensus GUAGUUUUUGCAAGAGGAAGAGAGCAGGAAGUGAAUGAGAAUUCUCUUUCCUUUUGUUUUGCGGCGAAGCGAUCCGCAAAGCAAUAUGUCGAAUUCGCCGCGAACGGAUCGCUUUUGUUU ((.((....(((((((((((((((.................))))).))))))))))...)).))(((((((((((....((.................))...)))))))))))..... (-35.79 = -35.79 + -0.00)



| Location | 12,063,321 – 12,063,440 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -31.71 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.64 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12063321 119 - 27905053 AAACAAAAGCGAUCCGUUCGCGGCGAAUUCGACGUAUUGCU-UGCGGAUCGCUUCGCCGCAAAACAAAAGGAAAGAGAAUUCUCAUUCACUUCCUGCUCUCUUCCUCUUGCAAAAACUAC ......(((((((((((..((.(((.......)))...)).-.)))))))))))....((((......(((((.((((...................))))))))).))))......... ( -32.31) >DroSec_CAF1 35353 120 - 1 AAACAAAAGCGAUCCGUUCGCGGCGAAUUCGACAUAUAGCUUUGCGGAUCGCUUCGCCGCAAAACAAAAGGAAAGAGAAUUCUCAUUCACUUCCUGCUCUCUUCCUCUUGCAAAAACUAC ......(((((((((((..((.(((....)).).....))...)))))))))))....((((......(((((.((((...................))))))))).))))......... ( -30.11) >DroSim_CAF1 44835 120 - 1 AAACAAAAGCGAUCCGUUCGCGGCGAAUUCGACAUAUCGCUUUGCGGAUCGCUUCGCCGCAAAACAAAAGGAAAGAGAAUUCUCAUUCACUUCCUGCUCUCUUCCUCUUGCAAAAACUAC ......(((((((((((....(((((..........)))))..)))))))))))....((((......(((((.((((...................))))))))).))))......... ( -32.71) >consensus AAACAAAAGCGAUCCGUUCGCGGCGAAUUCGACAUAUAGCUUUGCGGAUCGCUUCGCCGCAAAACAAAAGGAAAGAGAAUUCUCAUUCACUUCCUGCUCUCUUCCUCUUGCAAAAACUAC ......(((((((((((....(((((..........)))))..)))))))))))....((((......(((((.((((...................))))))))).))))......... (-30.42 = -30.64 + 0.22)



| Location | 12,063,361 – 12,063,475 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.65 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -31.38 |
| Energy contribution | -31.60 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12063361 114 - 27905053 -----CCUUGAAAAGCGAGCAAACCCAAAGUGGCAGCAAUAAACAAAAGCGAUCCGUUCGCGGCGAAUUCGACGUAUUGCU-UGCGGAUCGCUUCGCCGCAAAACAAAAGGAAAGAGAAU -----((((.....(((.((....((.....)).............(((((((((((..((.(((.......)))...)).-.))))))))))).))))).......))))......... ( -33.20) >DroSec_CAF1 35393 115 - 1 -----CCUUGAAAAGCGAGCAAGCCCAAAGUGGCAGCAAUAAACAAAAGCGAUCCGUUCGCGGCGAAUUCGACAUAUAGCUUUGCGGAUCGCUUCGCCGCAAAACAAAAGGAAAGAGAAU -----((((.....(((.((..(((......)))............(((((((((((..((.(((....)).).....))...))))))))))).))))).......))))......... ( -34.80) >DroSim_CAF1 44875 120 - 1 GAUUGCCUUGAAAAGCGAGCAAACCCAAAGUGGCAGCAAUAAACAAAAGCGAUCCGUUCGCGGCGAAUUCGACAUAUCGCUUUGCGGAUCGCUUCGCCGCAAAACAAAAGGAAAGAGAAU ..((((.(((.....))))))).......((((((....)......(((((((((((....(((((..........)))))..))))))))))).))))).................... ( -34.30) >consensus _____CCUUGAAAAGCGAGCAAACCCAAAGUGGCAGCAAUAAACAAAAGCGAUCCGUUCGCGGCGAAUUCGACAUAUAGCUUUGCGGAUCGCUUCGCCGCAAAACAAAAGGAAAGAGAAU .....((((.....(((.((....((.....)).............(((((((((((....(((((..........)))))..))))))))))).))))).......))))......... (-31.38 = -31.60 + 0.22)



| Location | 12,063,370 – 12,063,475 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -30.28 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12063370 105 - 27905053 --------------CCUUGAAAAGCGAGCAAACCCAAAGUGGCAGCAAUAAACAAAAGCGAUCCGUUCGCGGCGAAUUCGACGUAUUGCU-UGCGGAUCGCUUCGCCGCAAAACAAAAGG --------------((((.....(((.((....((.....)).............(((((((((((..((.(((.......)))...)).-.))))))))))).))))).......)))) ( -32.10) >DroSec_CAF1 35402 106 - 1 --------------CCUUGAAAAGCGAGCAAGCCCAAAGUGGCAGCAAUAAACAAAAGCGAUCCGUUCGCGGCGAAUUCGACAUAUAGCUUUGCGGAUCGCUUCGCCGCAAAACAAAAGG --------------((((.....(((.((..(((......)))............(((((((((((..((.(((....)).).....))...))))))))))).))))).......)))) ( -33.70) >DroSim_CAF1 44884 120 - 1 AUUGCACCUGAUUGCCUUGAAAAGCGAGCAAACCCAAAGUGGCAGCAAUAAACAAAAGCGAUCCGUUCGCGGCGAAUUCGACAUAUCGCUUUGCGGAUCGCUUCGCCGCAAAACAAAAGG ......(((..((((.(((.....))))))).......((((((....)......(((((((((((....(((((..........)))))..))))))))))).)))))........))) ( -36.10) >consensus ______________CCUUGAAAAGCGAGCAAACCCAAAGUGGCAGCAAUAAACAAAAGCGAUCCGUUCGCGGCGAAUUCGACAUAUAGCUUUGCGGAUCGCUUCGCCGCAAAACAAAAGG ..............((((.....(((.((....((.....)).............(((((((((((....(((((..........)))))..))))))))))).))))).......)))) (-30.28 = -30.50 + 0.22)


| Location | 12,063,475 – 12,063,594 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.12 |
| Mean single sequence MFE | -39.44 |
| Consensus MFE | -28.82 |
| Energy contribution | -29.46 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12063475 119 - 27905053 UCAUUUCCGCUUUGGG-CCGGAGUUUGUUGUUGCUUCGGGGCAGCAACAAUAACAACAGCAACUCUGCCACAAAGGCGACCAUUCGGAACCGCCGAGCGAGAAACUAUUGCACCUGAUUG ...(((((((((((((-(.((((((((((((((((....))))))))))...........))))))))).))..((((.((....))...))))))))).))))................ ( -39.10) >DroSec_CAF1 1 119 - 1 UCAUUUCCGCUUUAGG-CCGGAGUUGGUUGUUGCUUCGGGGCAGCAACAAUAACAACAGCAACUCUGCCACAAAGGCGACCAUUCGGAACCGCCGAGCGAGAAACUAUUGCACCUGAUUG ...(((((((((..((-(.((((((((((((((((....))))))))).....(....))))))))))).....((((.((....))...))))))))).))))................ ( -40.30) >DroSim_CAF1 1 105 - 1 UCAUUUCCGCUUUAGG-CCGGAGUUGGUUGUUGCUUCGGGGCAGCAACAAUAACAACAGCAACUCUGCCACAAAGGCGACCAUUCGGAACCGCCGAGCGAGAAACU-------------- ...(((((((((..((-(.((((((((((((((((....))))))))).....(....))))))))))).....((((.((....))...))))))))).))))..-------------- ( -40.30) >DroEre_CAF1 1985 119 - 1 UCAUUUCCGCUUUGGG-CCGGAGCUUGUUGUUGUUUCGGGGGAGCAACAAUAACAACAGCAACUCUGCCACAAAGCCGACCAUUCGGAACCGCCGAGCGAGAAACUAUUGCACCUGAUUG .....((.((((((((-(.((((.(((((((((((...(........)...)))))))))))))))))).)))))).))....((((.....))))((((.......))))......... ( -40.30) >DroYak_CAF1 2038 120 - 1 UCAUUUCCGCUUUGGGUCCGGUGCUUGUUGUUGUUUCAGGGGAGCAACAAUAAUAACAGCAACUCUGCCACAAAGGCGACCAUUCGGAACCGCCGAGCGAGAAACUAUUGCACCUGAUUG ...(((((((((.(((((((((((((((((((((((.....))))))))))).....))))....((((.....)))).....)))).))).).))))).))))................ ( -37.20) >consensus UCAUUUCCGCUUUGGG_CCGGAGUUUGUUGUUGCUUCGGGGCAGCAACAAUAACAACAGCAACUCUGCCACAAAGGCGACCAUUCGGAACCGCCGAGCGAGAAACUAUUGCACCUGAUUG ...(((((((((..((..(((((.(((((((((.....(........).....)))))))))))))))).....((((.((....))...))))))))).))))................ (-28.82 = -29.46 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:21 2006