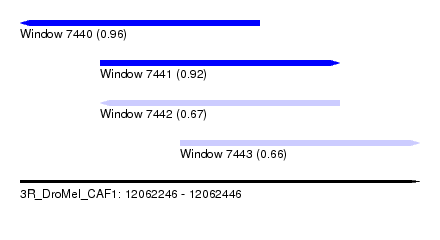
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,062,246 – 12,062,446 |
| Length | 200 |
| Max. P | 0.963320 |

| Location | 12,062,246 – 12,062,366 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -45.77 |
| Consensus MFE | -39.48 |
| Energy contribution | -41.57 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
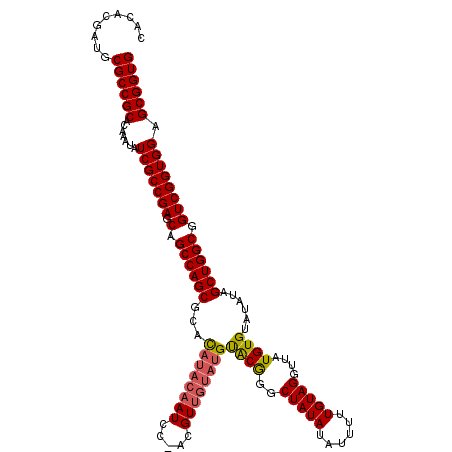
>3R_DroMel_CAF1 12062246 120 - 27905053 AAAAAUAUAUAGCCCGUACAUACAACGUGGGAUUGUAUGUGCGCUGGCUGCUCGGCGAUAUUUGUGCGGCGCAUCGUGUGCGCUGUCAGAGGCAAAUGGCUUGAUGGCCGUACACUCGUU .........(((((((((((((((((....).)))))))))))..)))))...(((((..((((.(((((((((...))))))))))))).....((((((....))))))....))))) ( -44.80) >DroSec_CAF1 34292 106 - 1 AAAAAUAUAUAGCCUGCGC--------------UAUAUAUGCGCUGGCUGCUCGGCGAUAUUUGUGCGGCGCAUCGUGUGCGCUGUCAGAGGCAAAUGGCUUGAUGGCCGUACAGUCGAU .........(((((.((((--------------.......)))).))))).((((((...((((.(((((((((...)))))))))))))..)..((((((....))))))...))))). ( -44.00) >DroSim_CAF1 43761 120 - 1 AAAAAUAUAUAGCCCGUACAUACAACGUCGGAUUGUAUGUGCGCUGGCUGCUCGGCGAUAUUUGUGCGGCGCAUCGUGUGCGCUGUCAGAGGCAAAUGGCUUGAUGGCCGUACAGUCGAU .........(((((((((((((((((....).)))))))))))..))))).((((((...((((.(((((((((...)))))))))))))..)..((((((....))))))...))))). ( -48.50) >consensus AAAAAUAUAUAGCCCGUACAUACAACGU_GGAUUGUAUGUGCGCUGGCUGCUCGGCGAUAUUUGUGCGGCGCAUCGUGUGCGCUGUCAGAGGCAAAUGGCUUGAUGGCCGUACAGUCGAU .........((((((((((((((((.......)))))))))))..))))).((((((...((((.(((((((((...)))))))))))))..)..((((((....))))))...))))). (-39.48 = -41.57 + 2.09)


| Location | 12,062,286 – 12,062,406 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -44.80 |
| Consensus MFE | -36.48 |
| Energy contribution | -37.43 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
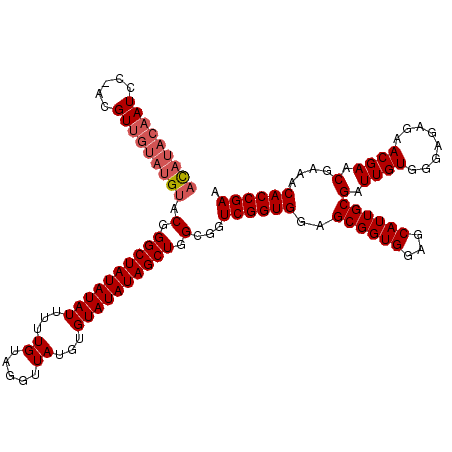
>3R_DroMel_CAF1 12062286 120 + 27905053 CACACGAUGCGCCGCACAAAUAUCGCCGAGCAGCCAGCGCACAUACAAUCCCACGUUGUAUGUACGGGCUAUAUAUUUUUGUAGGUUAUGUGUAUAUAGCUGGCUGUCGGUGGAGCGGUG .........((((((.......(((((((.(((((..((.(((((((((.....))))))))).))((((((((((...((.....))...)))))))))))))))))))))).)))))) ( -47.90) >DroSec_CAF1 34332 106 + 1 CACACGAUGCGCCGCACAAAUAUCGCCGAGCAGCCAGCGCAUAUAUA--------------GCGCAGGCUAUAUAUUUUUGUAGGUUAUGUGUAUAUAGCUGGCGGUCGGUGGAGCGGUG .........((((((.......(((((((.(.((((((..(((((((--------------.((.((.(((((......))))).)).))))))))).))))))).))))))).)))))) ( -41.60) >DroSim_CAF1 43801 120 + 1 CACACGAUGCGCCGCACAAAUAUCGCCGAGCAGCCAGCGCACAUACAAUCCGACGUUGUAUGUACGGGCUAUAUAUUUUUGUAGGUUAUGUGUAUAUAGCUGGCGGUCGGUGGAGCGGUG .........((((((.......(((((((.(.(((((((((((((...((((((((...)))).))))(((((......)))))..))))))).....))))))).))))))).)))))) ( -44.90) >consensus CACACGAUGCGCCGCACAAAUAUCGCCGAGCAGCCAGCGCACAUACAAUCC_ACGUUGUAUGUACGGGCUAUAUAUUUUUGUAGGUUAUGUGUAUAUAGCUGGCGGUCGGUGGAGCGGUG .........((((((.......(((((((.(.((((((...((((((((.....))))))))((((..(((((......)))))....))))......)))))).)))))))).)))))) (-36.48 = -37.43 + 0.95)


| Location | 12,062,286 – 12,062,406 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -30.11 |
| Energy contribution | -31.53 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12062286 120 - 27905053 CACCGCUCCACCGACAGCCAGCUAUAUACACAUAACCUACAAAAAUAUAUAGCCCGUACAUACAACGUGGGAUUGUAUGUGCGCUGGCUGCUCGGCGAUAUUUGUGCGGCGCAUCGUGUG ..((((.((.((((((((((((((((((.................)))))))).((((((((((((....).))))))))))).)))))).)))).)......).))))(((.....))) ( -40.43) >DroSec_CAF1 34332 106 - 1 CACCGCUCCACCGACCGCCAGCUAUAUACACAUAACCUACAAAAAUAUAUAGCCUGCGC--------------UAUAUAUGCGCUGGCUGCUCGGCGAUAUUUGUGCGGCGCAUCGUGUG ........((((((.((((.((((((((.................))))))))..((((--------------...((((.((((((....))))))))))..))))))))..))).))) ( -32.93) >DroSim_CAF1 43801 120 - 1 CACCGCUCCACCGACCGCCAGCUAUAUACACAUAACCUACAAAAAUAUAUAGCCCGUACAUACAACGUCGGAUUGUAUGUGCGCUGGCUGCUCGGCGAUAUUUGUGCGGCGCAUCGUGUG ..((((.((.(((((.((((((((((((.................)))))))).((((((((((((....).))))))))))).)))).).)))).)......).))))(((.....))) ( -35.83) >consensus CACCGCUCCACCGACCGCCAGCUAUAUACACAUAACCUACAAAAAUAUAUAGCCCGUACAUACAACGU_GGAUUGUAUGUGCGCUGGCUGCUCGGCGAUAUUUGUGCGGCGCAUCGUGUG ..((((.((.(((((.((((((((((((.................)))))))).(((((((((((.......))))))))))).)))).).)))).)......).))))(((.....))) (-30.11 = -31.53 + 1.42)



| Location | 12,062,326 – 12,062,446 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -28.75 |
| Energy contribution | -30.70 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12062326 120 + 27905053 ACAUACAAUCCCACGUUGUAUGUACGGGCUAUAUAUUUUUGUAGGUUAUGUGUAUAUAGCUGGCUGUCGGUGGAGCGGUGGAGCAUUGCGAUUGUGGGAGAGAACGAACGAAACACCGAA (((((((((.....))))))))).(.((((((((((...((.....))...)))))))))).)...((((((..((((((...))))))(.((((........)))).)....)))))). ( -36.20) >DroSec_CAF1 34372 106 + 1 AUAUAUA--------------GCGCAGGCUAUAUAUUUUUGUAGGUUAUGUGUAUAUAGCUGGCGGUCGGUGGAGCGGUGGAGCAUUGCGAUUGUGGGAGAGAACGAACGAAACACCGAA .......--------------.(((.((((((((((...((.....))...)))))))))).))).((((((..((((((...))))))(.((((........)))).)....)))))). ( -32.50) >DroSim_CAF1 43841 120 + 1 ACAUACAAUCCGACGUUGUAUGUACGGGCUAUAUAUUUUUGUAGGUUAUGUGUAUAUAGCUGGCGGUCGGUGGAGCGGUGGAGCAUUGCGAUUGUGGGAGAGAACGAACGAAACACCGAA (((((((((.....))))))))).(.((((((((((...((.....))...)))))))))).)...((((((..((((((...))))))(.((((........)))).)....)))))). ( -36.20) >consensus ACAUACAAUCC_ACGUUGUAUGUACGGGCUAUAUAUUUUUGUAGGUUAUGUGUAUAUAGCUGGCGGUCGGUGGAGCGGUGGAGCAUUGCGAUUGUGGGAGAGAACGAACGAAACACCGAA (((((((((.....))))))))).(.((((((((((...((.....))...)))))))))).)...((((((..((((((...))))))(.((((........)))).)....)))))). (-28.75 = -30.70 + 1.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:16 2006