
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,050,348 – 12,050,461 |
| Length | 113 |
| Max. P | 0.971804 |

| Location | 12,050,348 – 12,050,461 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.53 |
| Mean single sequence MFE | -41.61 |
| Consensus MFE | -32.83 |
| Energy contribution | -33.50 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.89 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
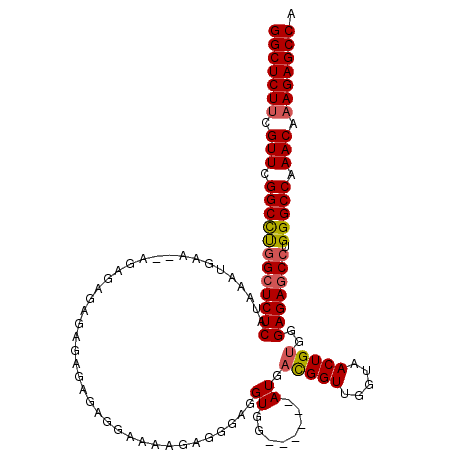
>3R_DroMel_CAF1 12050348 113 + 27905053 GGCUCUUCGUUCGGCCUGGCUCUCAUAAAUGACAUAGAGAGAGAGAGAGAGGAAAAGAGGGAGGUGG------AUGACGGUUGGUAACUGUGGGAGAGCGUGGGCCAAACAAAGAGCCA (((((((.(((.((((((((((((........(((............................))).------.(.(((((.....))))).))))))).)))))).))).))))))). ( -38.29) >DroSim_CAF1 21709 111 + 1 GGCUCUUCGUUCGGCCUGGCUCUCAUAAAUGAA--CGAGAGAGAGAGAGAGGAAAAGAGGGAGGUGG------AUGACGGUUGGUAACUGUGGGAGAGCCUGGGCCAAACAAAGAGCCA (((((((.(((.((((((((((((.........--(....)..........................------.(.(((((.....))))).)))))))).))))).))).))))))). ( -43.10) >DroYak_CAF1 21440 117 + 1 GGCUCUCCGUUCGGCUCGGCUCUCAUGAAAGAG--AGAGAGAGAGCGAGUGGAAGGGAGGGAGGUGGAGGUCGAUGUUGGUUGGUAACUGUGGGAGAACCUGGGCCAAACAAAGAGCCA ((((((((.(((.(((((.(((((.........--.....)))))))))).))).)))....(((..(((((.....((((.....)))).....).))))..))).......))))). ( -43.44) >consensus GGCUCUUCGUUCGGCCUGGCUCUCAUAAAUGAA__AGAGAGAGAGAGAGAGGAAAAGAGGGAGGUGG______AUGACGGUUGGUAACUGUGGGAGAGCCUGGGCCAAACAAAGAGCCA (((((((.(((.((((((((((((.......................................((........)).(((((.....)))))..))))))).))))).))).))))))). (-32.83 = -33.50 + 0.67)

| Location | 12,050,348 – 12,050,461 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.53 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -33.16 |
| Energy contribution | -33.05 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -5.73 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12050348 113 - 27905053 UGGCUCUUUGUUUGGCCCACGCUCUCCCACAGUUACCAACCGUCAU------CCACCUCCCUCUUUUCCUCUCUCUCUCUCUCUAUGUCAUUUAUGAGAGCCAGGCCGAACGAAGAGCC .((((((((((((((((...((((((..((.((.....)).))...------................................(((.....)))))))))..)))))))))))))))) ( -35.60) >DroSim_CAF1 21709 111 - 1 UGGCUCUUUGUUUGGCCCAGGCUCUCCCACAGUUACCAACCGUCAU------CCACCUCCCUCUUUUCCUCUCUCUCUCUCUCG--UUCAUUUAUGAGAGCCAGGCCGAACGAAGAGCC .((((((((((((((((..(((((((....................------................................--.........))))))).)))))))))))))))) ( -38.65) >DroYak_CAF1 21440 117 - 1 UGGCUCUUUGUUUGGCCCAGGUUCUCCCACAGUUACCAACCAACAUCGACCUCCACCUCCCUCCCUUCCACUCGCUCUCUCUCU--CUCUUUCAUGAGAGCCGAGCCGAACGGAGAGCC .(((((((((((((((.(.(((.((.....))..)))....................................((((((.....--.........)))))).).))))))))))))))) ( -36.74) >consensus UGGCUCUUUGUUUGGCCCAGGCUCUCCCACAGUUACCAACCGUCAU______CCACCUCCCUCUUUUCCUCUCUCUCUCUCUCU__CUCAUUUAUGAGAGCCAGGCCGAACGAAGAGCC .(((((((((((((((...(((((((.....................................................................)))))))..))))))))))))))) (-33.16 = -33.05 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:03 2006