
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,044,691 – 12,044,784 |
| Length | 93 |
| Max. P | 0.983731 |

| Location | 12,044,691 – 12,044,784 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.60 |
| Mean single sequence MFE | -27.89 |
| Consensus MFE | -18.87 |
| Energy contribution | -17.90 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12044691 93 - 27905053 UCCGCUAAUUAGUAAUACCGCCGAAGUGGUGAACUCUACUUAACGUCAUUCU---------------------------UGUUGCGGCUUUUUAGCCGCAGAAACUAAGGCAAACCUGAU ...(((..(((((..((((((....)))))).....................---------------------------..((((((((....))))))))..))))))))......... ( -25.50) >DroSec_CAF1 15919 93 - 1 UCCGCUAAUUAGCAAUGCCGCCAAAGUGGUGAACUCUACUUAACGCCAUUCU---------------------------UGUUGUGGCUUUUUAGCCGCAGAAACUAAGGCAAACCUUAU ...(((....)))..((((..((((((((((............))))))).)---------------------------))((((((((....)))))))).......))))........ ( -26.90) >DroSim_CAF1 15913 93 - 1 UCCGCUAAUUAGCAAUGCCGCCAAAGUGGUGAACUCUACUUAACGCCAUUCU---------------------------UGUUGUGGCUUUUUAGCCGCAGAAACUAAGGCAAACCUGAU ...(((....)))..((((..((((((((((............))))))).)---------------------------))((((((((....)))))))).......))))........ ( -26.90) >DroEre_CAF1 15235 93 - 1 UCAGCUAAUUAGCAAUACCGCCAAAGUGGUGAACUCUACUUAACGGCAUUCU---------------------------AAUUGUGGCUUUUUAGCCGCAAAAACUAAGACAAACCUGAU ((((..((((((.(((.(((...((((((......))))))..))).)))))---------------------------))))((((((....))))))................)))). ( -25.70) >DroYak_CAF1 15511 93 - 1 UCGGCUAAUUAGCAAUACCGCCAAUGUGGCGAACUCCACUUAACGCCAUUCU---------------------------UGUUGUGGCUUUUUAGCCGCAGAAACUAAGACAAACCUGAU .(((((((..(((..(((.((....((((((............))))))...---------------------------.)).))))))..)))))))(((..............))).. ( -24.74) >DroAna_CAF1 14575 120 - 1 AUGCCUAAUUAGGGGCAACACCAAAGUGGUAAGUCCUACUUGGCCCUAGUCCGUGUGUAGGUAGUGCUCCUGCUCCUGCUCCUGCGGUCGCUUAACCGCAGAAAGUAAGACAAACCUGAU .(((((......)))))..........(((..(((.(((((...............(((((.(((......))))))))..(((((((......))))))).))))).)))..))).... ( -37.60) >consensus UCCGCUAAUUAGCAAUACCGCCAAAGUGGUGAACUCUACUUAACGCCAUUCU___________________________UGUUGUGGCUUUUUAGCCGCAGAAACUAAGACAAACCUGAU ...(((....)))..........((((((......))))))........................................((((((((....))))))))................... (-18.87 = -17.90 + -0.97)

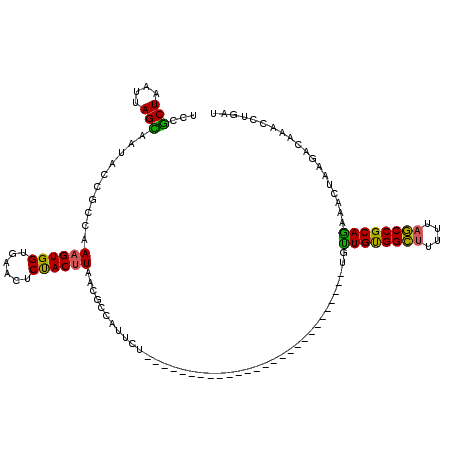
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:55 2006