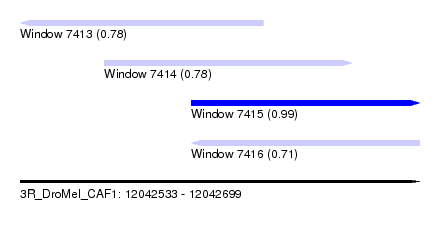
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,042,533 – 12,042,699 |
| Length | 166 |
| Max. P | 0.986419 |

| Location | 12,042,533 – 12,042,634 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -31.96 |
| Energy contribution | -32.40 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12042533 101 - 27905053 GUGCUGCUGUACAUGUGUGCGUGUGUGU--GUGUACUGUUUGUUAGUAUGUAUUUGGGUCACAUGCAUACACGCACGGGGCCACAGACAGCCGCCAACAAGGA (.((((((((...(.(((((((((((((--(((((((....((........))...))).))))))))))))))))).)...)))).))))).((.....)). ( -38.80) >DroSec_CAF1 13867 103 - 1 GUGCUGCUGUACAUGUGUGCGAGUGUGUGGGUGUGCUGUUUGUUAGUAUGUAUUUGGGUCACAUGCAUACACGCACGGGGCCACAGACAGCCGCCAACAAGGA .....(((((((....)))).))).(((.((((.(((((((((..((((((((.((...)).))))))))..((.....)).))))))))))))).))).... ( -35.10) >DroSim_CAF1 13772 101 - 1 GUGCUGCUGUACAUGUGUGCGAGUGUGU--GUGUGCUGUUUGUUAGUAUGUAUUUGGGUCAUAUGCAUACGCGCACGGGGCCACAGACAGCCGCCAACAAGGA (.((.(((((...((((.((..((((((--((((((.........(((((.........)))))))))))))))))...)))))).))))).)))........ ( -38.10) >DroYak_CAF1 13333 101 - 1 GUGCUGCUGUACAUGUGUGUUUGCGUGU--GUGUGCUGUUUGUUAGUAUGUAUUUGGGUCACAUGCAUACACGCACGGGGCCACAGACAGCCGCCAACAAGGA (.((.(((((...((((.((((.(((((--((((((((.....))))((((((.((...)).))))))))))))))))))))))).))))).)))........ ( -36.50) >consensus GUGCUGCUGUACAUGUGUGCGAGUGUGU__GUGUGCUGUUUGUUAGUAUGUAUUUGGGUCACAUGCAUACACGCACGGGGCCACAGACAGCCGCCAACAAGGA (.((((((((...(.(((((((((((((..(..(((((.....)))))..)...((.....)).))))))))))))).)...)))).))))).((.....)). (-31.96 = -32.40 + 0.44)

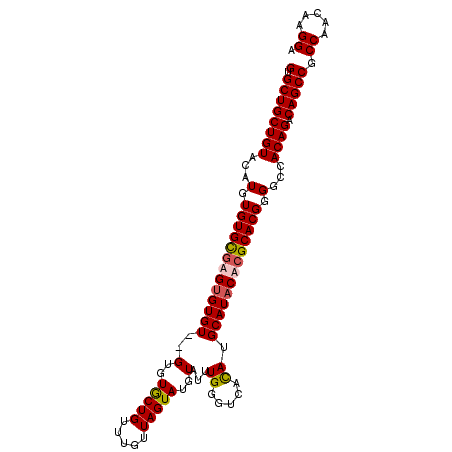

| Location | 12,042,568 – 12,042,671 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.99 |
| Mean single sequence MFE | -12.93 |
| Consensus MFE | -11.24 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12042568 103 + 27905053 AUGCAUGUGACCCAAAUACAUACUAACAAACAGUACAC--ACACACACGCACACAUGUACAGCAGCACCAACACACAAUGGCUGUUCAUUCAUAUAAGCCAAACA .((((((((...........((((.......))))...--...........))))))))...................(((((.............))))).... ( -13.83) >DroSec_CAF1 13902 105 + 1 AUGCAUGUGACCCAAAUACAUACUAACAAACAGCACACCCACACACUCGCACACAUGUACAGCAGCACCAACACACAAUGGCUGUUCAUUCAUAUAAGCCAAACA .((((((((.......((.....)).......((..............)).))))))))...................(((((.............))))).... ( -12.96) >DroSim_CAF1 13807 103 + 1 AUGCAUAUGACCCAAAUACAUACUAACAAACAGCACAC--ACACACUCGCACACAUGUACAGCAGCACCAACACACAAUGGCUGUUCAUUCAUAUAAGCCAAACA ..((((((((......................((....--........))....(((.(((((.................))))).)))))))))..))...... ( -10.73) >DroYak_CAF1 13368 103 + 1 AUGCAUGUGACCCAAAUACAUACUAACAAACAGCACAC--ACACGCAAACACACAUGUACAGCAGCACCAACACACAAUGGCUGUUCAUUCAUAUAAGCCAAACA .((((((((.......((.....)).......((....--....)).....))))))))...................(((((.............))))).... ( -14.22) >consensus AUGCAUGUGACCCAAAUACAUACUAACAAACAGCACAC__ACACACACGCACACAUGUACAGCAGCACCAACACACAAUGGCUGUUCAUUCAUAUAAGCCAAACA .((((((((..........................................))))))))...................(((((.............))))).... (-11.24 = -11.50 + 0.25)



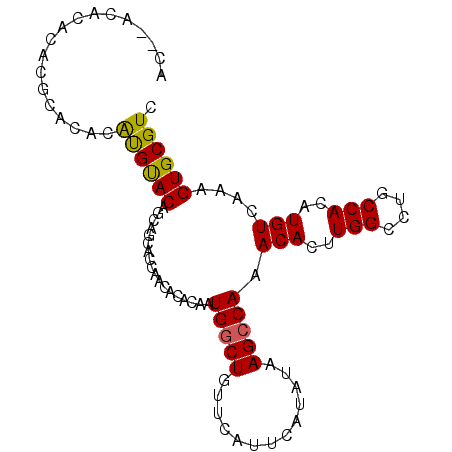
| Location | 12,042,604 – 12,042,699 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 85.70 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -15.58 |
| Energy contribution | -15.30 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12042604 95 + 27905053 AC--ACACACACGCACACAUGUACAGCAGCACCAACACACAAUGGCUGUUCAUUCAUAUAAGCCAAACACUUGGCCUGCCACAUGUCAAAGUGCGUC ..--......((((((((((((...((((..((((.......(((((.............))))).....)))).)))).))))))....)))))). ( -26.52) >DroSec_CAF1 13938 97 + 1 ACCCACACACUCGCACACAUGUACAGCAGCACCAACACACAAUGGCUGUUCAUUCAUAUAAGCCAAACACUUGGCCUGCCACAUGUCAAAGUGCGUC ...........(((((((((((...((((..((((.......(((((.............))))).....)))).)))).))))))....))))).. ( -25.12) >DroSim_CAF1 13843 95 + 1 AC--ACACACUCGCACACAUGUACAGCAGCACCAACACACAAUGGCUGUUCAUUCAUAUAAGCCAAACACUUGGCCUGCCACAUGUCAAAGUGCGUC ..--.......(((((((((((...((((..((((.......(((((.............))))).....)))).)))).))))))....))))).. ( -25.12) >DroYak_CAF1 13404 95 + 1 AC--ACACGCAAACACACAUGUACAGCAGCACCAACACACAAUGGCUGUUCAUUCAUAUAAGCCAAACACUUGGCCUGCCACUUGUCAAAGUGCGUC ..--..(((((.....(((.((...((((..((((.......(((((.............))))).....)))).)))).)).))).....))))). ( -21.32) >DroAna_CAF1 12489 72 + 1 ------------------GCGCACAGC-------ACACACAAUGCCUGUUCAUUCAUAUGAGCCAAACACUUGGCCUGCCACAUGUCAAAGUGCGUC ------------------((((((.((-------(.......)))..((((((....))))))...(((..(((....)))..)))....)))))). ( -18.70) >consensus AC__ACACACACGCACACAUGUACAGCAGCACCAACACACAAUGGCUGUUCAUUCAUAUAAGCCAAACACUUGGCCUGCCACAUGUCAAAGUGCGUC ..................((((((..................(((((.............))))).(((..(((....)))..)))....)))))). (-15.58 = -15.30 + -0.28)


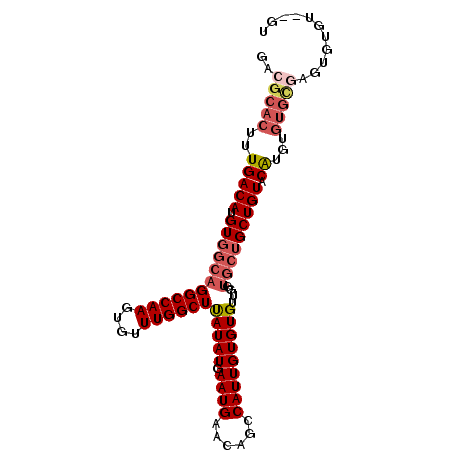
| Location | 12,042,604 – 12,042,699 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.70 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -20.32 |
| Energy contribution | -21.88 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12042604 95 - 27905053 GACGCACUUUGACAUGUGGCAGGCCAAGUGUUUGGCUUAUAUGAAUGAACAGCCAUUGUGUGUUGGUGCUGCUGUACAUGUGUGCGUGUGUGU--GU .((((((....(((((((((((((((((..(.(((((.............)))))..)..).))))).)))))..))))))))))))......--.. ( -31.32) >DroSec_CAF1 13938 97 - 1 GACGCACUUUGACAUGUGGCAGGCCAAGUGUUUGGCUUAUAUGAAUGAACAGCCAUUGUGUGUUGGUGCUGCUGUACAUGUGUGCGAGUGUGUGGGU .((((((((..(((((((((((((((((..(.(((((.............)))))..)..).))))).)))))..))))))....)))))))).... ( -32.52) >DroSim_CAF1 13843 95 - 1 GACGCACUUUGACAUGUGGCAGGCCAAGUGUUUGGCUUAUAUGAAUGAACAGCCAUUGUGUGUUGGUGCUGCUGUACAUGUGUGCGAGUGUGU--GU .((((((((..(((((((((((((((((..(.(((((.............)))))..)..).))))).)))))..))))))....))))))))--.. ( -32.52) >DroYak_CAF1 13404 95 - 1 GACGCACUUUGACAAGUGGCAGGCCAAGUGUUUGGCUUAUAUGAAUGAACAGCCAUUGUGUGUUGGUGCUGCUGUACAUGUGUGUUUGCGUGU--GU ...(((((...((((((((((((((((....)))))))...((......)))))))).)))...))))).....(((((((......))))))--). ( -29.80) >DroAna_CAF1 12489 72 - 1 GACGCACUUUGACAUGUGGCAGGCCAAGUGUUUGGCUCAUAUGAAUGAACAGGCAUUGUGUGU-------GCUGUGCGC------------------ ..(((((...(((((.(((....))).))))).(((.(((((.((((......))))))))).-------)))))))).------------------ ( -24.20) >consensus GACGCACUUUGACAUGUGGCAGGCCAAGUGUUUGGCUUAUAUGAAUGAACAGCCAUUGUGUGUUGGUGCUGCUGUACAUGUGUGCGAGUGUGU__GU ..(((((..(((((.((((((((((((....))))))(((((.((((......)))))))))....))))))))).))...)))))........... (-20.32 = -21.88 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:51 2006