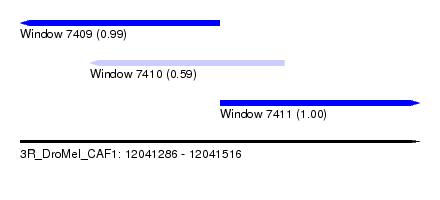
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,041,286 – 12,041,516 |
| Length | 230 |
| Max. P | 0.996348 |

| Location | 12,041,286 – 12,041,401 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.29 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -19.76 |
| Energy contribution | -21.36 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12041286 115 - 27905053 UGGAAUAUAUUUUCGGGCUCUGUGCUGCUACCAGCACGAGUCCUGCAACCCCAAGUCCUUUCCUUGGCC----CCA-UCCCAGCUCGACCUAACAUCAAUUCCUUCCACCGACCGCCUCU .(((((........((((((.((((((....)))))))))))).......(((((.......)))))..----...-.....................)))))................. ( -27.90) >DroSec_CAF1 12662 115 - 1 UGGAAUAUAUUUUCGGGCUCUGUGCUGCUACCAGCACGAGUCCUGCAACCCCAAGUCCUUUCCUUGGCC----CCA-UCCCAGCUCGACCUAACAUCAAUUCCUUCCACCGACCACCUCU .(((((........((((((.((((((....)))))))))))).......(((((.......)))))..----...-.....................)))))................. ( -27.90) >DroEre_CAF1 11865 115 - 1 UGGAAUAUAUUUUCGGUCUCUGUGCUGCUACCAGCACGAGUCCUGCCACCCCAAGUCCUUUCCUUUGCC----CCA-UCCCAGCUCGACCUAACAUCAAUUCCUUCCACCGACCUCCUCU .(((((........((((...((((((....))))))((((..........((((........))))..----...-.....))))))))........)))))................. ( -22.04) >DroYak_CAF1 12142 115 - 1 UGGAAUAUAUUUUCGGUCUCUGUGCUGCUACCAGCACGAGUCCUGCAACCCCAAGUCCUUUCCUUGGCC----CCA-UCCCAGCUCGACCUAACAUCAAUUCCUUCCACCGACCCCCUCA .(((((........((((...((((((....))))))((((.........(((((.......)))))..----...-.....))))))))........)))))................. ( -26.80) >DroAna_CAF1 11209 104 - 1 UGGAAUAUAUUUC--GGCUCUGUGCUGCUACCAGCAAGAGUCCUCCAACCCCAGGUCCUUU---UUGCCCAUUCCAUUCCAAGCUCAACCUAACAUCAAUUCCUUCUAC----------- ((((((.......--((((((.(((((....)))))))))))...........(((.....---..)))......))))))............................----------- ( -20.50) >consensus UGGAAUAUAUUUUCGGGCUCUGUGCUGCUACCAGCACGAGUCCUGCAACCCCAAGUCCUUUCCUUGGCC____CCA_UCCCAGCUCGACCUAACAUCAAUUCCUUCCACCGACC_CCUCU .(((((........((((((.((((((....)))))))))))).......(((((.......)))))...............................)))))................. (-19.76 = -21.36 + 1.60)

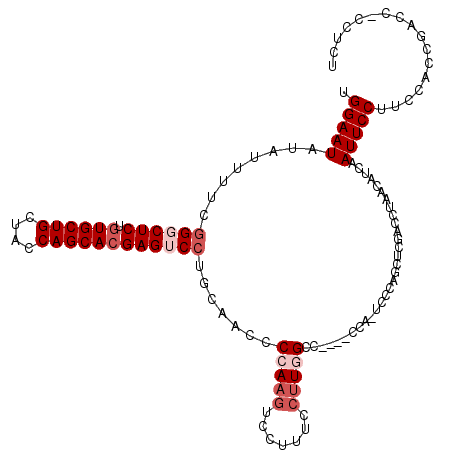
| Location | 12,041,326 – 12,041,438 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -18.56 |
| Energy contribution | -19.96 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12041326 112 - 27905053 CUAGCUUAAAUGCCAGAAAUUUAAUAAACAAUUCUAGUGGAAUAUAUUUUCGGGCUCUGUGCUGCUACCAGCACGAGUCCUGCAACCCCAAGUCCUUUCCUUGGCC----CCA-UCC ((.((......)).))....................((((...........((((((.((((((....)))))))))))).......(((((.......)))))..----)))-).. ( -28.70) >DroSec_CAF1 12702 112 - 1 CCAGCUUAAAUGCCAGAAAUUUAAUAAACAAUUCUAGUGGAAUAUAUUUUCGGGCUCUGUGCUGCUACCAGCACGAGUCCUGCAACCCCAAGUCCUUUCCUUGGCC----CCA-UCC ...((......)).......................((((...........((((((.((((((....)))))))))))).......(((((.......)))))..----)))-).. ( -27.60) >DroEre_CAF1 11905 112 - 1 CCAGCUUAAAUGCCAGAAAUUUAAUAAACAAUUCUAGUGGAAUAUAUUUUCGGUCUCUGUGCUGCUACCAGCACGAGUCCUGCCACCCCAAGUCCUUUCCUUUGCC----CCA-UCC ...((((((((.......))))))............((((((......)))((.(((.((((((....))))))))).))...))).................)).----...-... ( -20.40) >DroYak_CAF1 12182 112 - 1 CCAGCUUAAAUGCCAGAAAUUUAAUAAACAAUUCUAGUGGAAUAUAUUUUCGGUCUCUGUGCUGCUACCAGCACGAGUCCUGCAACCCCAAGUCCUUUCCUUGGCC----CCA-UCC ...((......)).......................((((...........((.(((.((((((....))))))))).)).......(((((.......)))))..----)))-).. ( -24.30) >DroAna_CAF1 11238 111 - 1 -UGGCUUAAAUGCCAGAAAUUUAAUAAACAAUUCUAGUGGAAUAUAUUUC--GGCUCUGUGCUGCUACCAGCAAGAGUCCUCCAACCCCAGGUCCUUU---UUGCCCAUUCCAUUCC -((((......))))....................((((((((.......--((((((.(((((....)))))))))))...........(((.....---..))).)))))))).. ( -27.70) >consensus CCAGCUUAAAUGCCAGAAAUUUAAUAAACAAUUCUAGUGGAAUAUAUUUUCGGGCUCUGUGCUGCUACCAGCACGAGUCCUGCAACCCCAAGUCCUUUCCUUGGCC____CCA_UCC ...........(((((..............((((.....))))........((((((.((((((....))))))))))))....................)))))............ (-18.56 = -19.96 + 1.40)



| Location | 12,041,401 – 12,041,516 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.83 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -25.40 |
| Energy contribution | -25.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12041401 115 + 27905053 CUAGAAUUGUUUAUUAAAUUUCUGGCAUUUAAGCUAGAGCUGUUGUUUGGAAUUGG-GAAAACCGACGCUCUAGACAAAGCAAAGCACAAGAACAAAAUCAAAAUGCAAUCUAGCA (((((.((((..........((((((......))))))(((.(((((((((.((((-.....))))...))))))))))))........................))))))))).. ( -29.70) >DroSec_CAF1 12777 115 + 1 CUAGAAUUGUUUAUUAAAUUUCUGGCAUUUAAGCUGGAGCUGUUGUUUGGAAUUGG-GAAAACCGACGCUCUAGACAAAGCAAAGCACAAGAACAGAAUCAAAAUGCAAUCUAGCA (((((.((((.........(((((........(((...(((.(((((((((.((((-.....))))...))))))))))))..))).......))))).......))))))))).. ( -29.75) >DroEre_CAF1 11980 116 + 1 CUAGAAUUGUUUAUUAAAUUUCUGGCAUUUAAGCUGGAGCUGUUGUUUGGAAUCGGGGAAAACCGACGCUCUAGACAAAGCAAAGCUCAAGAACAGAAUCAAAAUGCAAUCUAGCA (((((.((((.........(((((((......))..(((((.(((((((((.((((......))))...))))))))).....))))).....))))).......))))))))).. ( -34.59) >DroYak_CAF1 12257 116 + 1 CUAGAAUUGUUUAUUAAAUUUCUGGCAUUUAAGCUGGAGCUGUUGUUUGGAAUUGGGGAAAACCGACGCUCUAAACAAAGCAAAGCACAAGAACAGAAUCAAAGUGCAAUCUAGCA ((((((.............)))))).......(((((((((.(((((((((.((((......))))...))))))))))))...((((..((......))...))))..)))))). ( -32.62) >DroAna_CAF1 11313 113 + 1 CUAGAAUUGUUUAUUAAAUUUCUGGCAUUUAAGCCA--GCUUGACGCUAGAAUUGGUGAAAACCAAAGCACCGGACAGAGCGAACCAGGGGAAUGGAAUCAAAUUGCAAUCUGGG- (((((.((((...........(((((......))))--).((((((((....((((((..........))))))....))))..(((......)))..))))...))))))))).- ( -29.80) >consensus CUAGAAUUGUUUAUUAAAUUUCUGGCAUUUAAGCUGGAGCUGUUGUUUGGAAUUGG_GAAAACCGACGCUCUAGACAAAGCAAAGCACAAGAACAGAAUCAAAAUGCAAUCUAGCA (((((.((((..........((((((......))))))(((.(((((((((.((((......))))...))))))))))))........................))))))))).. (-25.40 = -25.64 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:47 2006