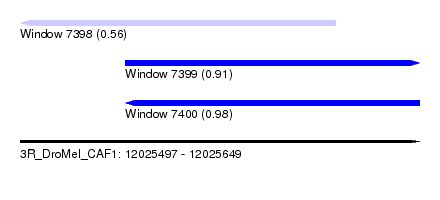
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,025,497 – 12,025,649 |
| Length | 152 |
| Max. P | 0.977162 |

| Location | 12,025,497 – 12,025,617 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -40.78 |
| Consensus MFE | -24.04 |
| Energy contribution | -23.43 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12025497 120 - 27905053 GGGGAGUUUGGAAAGUACCAGUUCCUGCAGUUCUUCCUGCAGGUGCUCUCCGGCCUGACCGCUGGCAUGCACAUGCUAUCGCUGGUCACGGUGGCCGCCGUGCCUGAGCAUCGCUGCUUC (((((((((((......))))..(((((((......))))))).))))))).((((((((((((((((....))))))..)).))))).)))(((.((.((((....)))).)).))).. ( -52.40) >DroVir_CAF1 14003 120 - 1 GGUGAAUUUGGUAAAUAUCAAUUUGCCCAAUUCUUUCUGCAAGUGCUGUCCGCUCUCACGGCGGGUAUGCACAUGCUCUCGCUGGUGACUGUUGCUGCUGUGCCCGAGCAUCGCUGCUUU ...(((((.(((((((....))))))).))))).....(((.(((((..(((((.....)))))..).)))).)))....((.(....).)).((.((.((((....)))).)).))... ( -40.10) >DroPse_CAF1 12122 120 - 1 GGGGAAUUUGGUAAAUACCAAUUCUUGCAGUUCUUCCUGCAAGUCCUCUCCGCUUUGACCGCAGGCAUGCAUAUGCUCUCCCUGGUUACGGUAUCCGCAGUGCCAGAGCAUCGCUGCUUU (((((..(((((....)))))..(((((((......)))))))...)))))((.......))(((((.((..(((((((....(((..(((...)))....)))))))))).))))))). ( -39.10) >DroGri_CAF1 4253 120 - 1 GGUGAAUUUGGCAAAUAUCAAUUUGCCCAAUUCUUUUUGCAAGUGCUCUCGGCACUGACGGCGGGAAUGCAUAUGCUGUCUCUGGUAACUGUGGCUGCUGUUCCGGAGCAUCGGUGCUUU ...(((((.(((((((....))))))).))))).....(((.((((((.(((....((((((((...((((..((((......))))..)))).)))))))))))))))))...)))... ( -41.90) >DroWil_CAF1 3659 120 - 1 GGGGAAUUUGGUAAAUACCAAUUCCUUCAAUUCUUCCUGCAAGUAUUAUCCGCCUUGACUGCUGGUCUGCAUAUGCUCUCACUGGUCACAGUUGCCGCUGUGCCCGAGCAUCGUUGUUUC ((((((((.(((....)))))))))))...........((((.........((...(((.....))).))..((((((.....((.((((((....)))))).))))))))..))))... ( -30.20) >DroMoj_CAF1 12540 120 - 1 GGUGAAUUCGGCAAAUACCAAUUCGCCCAAUUUUUCCUGCAAGUGUUGUCCGCAUUGACAGCGGGCAUGCACAUGCUGUCCCUGGUGACUGUUGCAUCAGUGCCCGAGCAUCGCUGCUUU ((((((((.((......))))))))))...........(((.(((((((((((.......))))))).))))((((((..(((((((.(....))))))).)..).)))))...)))... ( -41.00) >consensus GGGGAAUUUGGUAAAUACCAAUUCGCCCAAUUCUUCCUGCAAGUGCUCUCCGCACUGACCGCGGGCAUGCACAUGCUCUCCCUGGUCACUGUUGCCGCUGUGCCCGAGCAUCGCUGCUUU ((((((((.((......))))))))))...........(((.((((...((((.......)).))...)))).))).......((.((((((....)))))))).(((((....))))). (-24.04 = -23.43 + -0.61)

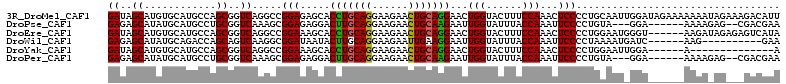
| Location | 12,025,537 – 12,025,649 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.77 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -18.13 |
| Energy contribution | -17.68 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12025537 112 + 27905053 GAUAGCAUGUGCAUGCCAGCGGUCAGGCCGGAGAGCACCUGCAGGAAGAACUGCAGGAACUGGUACUUUCCAAACUCCCCUGCAAUUGGAUAGAAAAAAAUAGAAAGACAUU ....((....))...((((..(.((((..((((....(((((((......)))))))...(((......)))..)))))))))..))))....................... ( -32.70) >DroPse_CAF1 12162 101 + 1 GAGAGCAUAUGCAUGCCUGCGGUCAAAGCGGAGAGGACUUGCAGGAAGAACUGCAAGAAUUGGUAUUUACCAAAUUCCCCUGUA---GGA------AAAAGAG--CGACGAA ....((....)).(((((....((...((((.(.((((((((((......)))))))..(((((....)))))..)))))))).---.))------...)).)--))..... ( -28.70) >DroEre_CAF1 3838 106 + 1 GAUAGCAUGUGCAUGCCAGCGGUCAGGCCGGAAAGCACCUGCAGGAAGAACUGCAGGAACUGGUACUUUCCAAACUCCCCUGGAAUGGGU------AAGAUAGAGAGUCAUA ....((....)).(((((((((.....))).......(((((((......)))))))..))))))(((((.......(((......))).------......)))))..... ( -34.44) >DroWil_CAF1 3699 96 + 1 GAGAGCAUAUGCAGACCAGCAGUCAAGGCGGAUAAUACUUGCAGGAAGAAUUGAAGGAAUUGGUAUUUACCAAAUUCCCCUAAAAUGAUC------AAG----------GAA ..((.(((.(((......)))....(((.(((.....(((.(((......))))))...(((((....)))))..))))))...))).))------...----------... ( -16.90) >DroYak_CAF1 3804 92 + 1 GAUAGCAUGUGCAUGCCAGCGGUCAGGCCGGAAAGCACCUGCAGGAAGAACUGCAGGAACUGGUACUUUCCAAACUCCCCUGGAAUUGGA------A--------------A ....((....)).(((((((((.....))).......(((((((......)))))))..))))))..((((((..(((...))).)))))------)--------------. ( -33.00) >DroPer_CAF1 12128 101 + 1 GAGAGCAUAUGCAUGCCUGCGGUCAAAGCGGAGAGGACUUGCAGGAAGAACUGCAAGAAUUGGUAUUUACCAAAUUCCCCUGUA---GGA------AAAAGAG--CGACGAA ....((....)).(((((....((...((((.(.((((((((((......)))))))..(((((....)))))..)))))))).---.))------...)).)--))..... ( -28.70) >consensus GAGAGCAUAUGCAUGCCAGCGGUCAAGCCGGAGAGCACCUGCAGGAAGAACUGCAGGAACUGGUACUUACCAAACUCCCCUGGAAU_GGA______AAAA_AG___GACGAA ((..((............))..)).....(((.....(((((((......)))))))...(((......)))...))).................................. (-18.13 = -17.68 + -0.44)

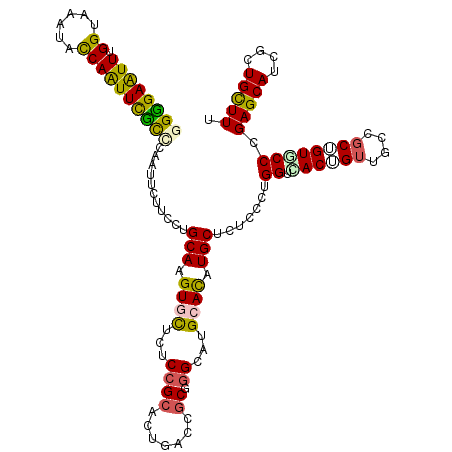
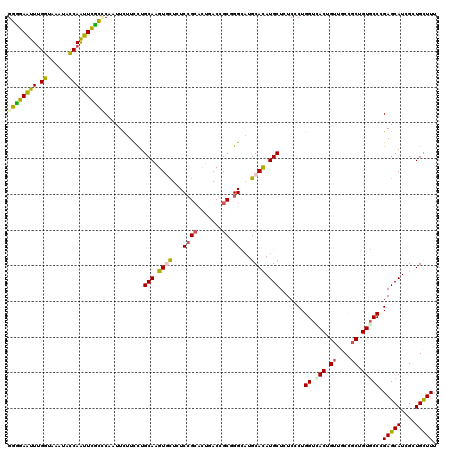
| Location | 12,025,537 – 12,025,649 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.77 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -20.99 |
| Energy contribution | -21.35 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
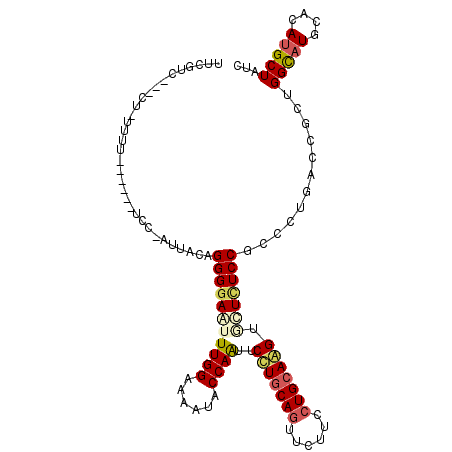
>3R_DroMel_CAF1 12025537 112 - 27905053 AAUGUCUUUCUAUUUUUUUCUAUCCAAUUGCAGGGGAGUUUGGAAAGUACCAGUUCCUGCAGUUCUUCCUGCAGGUGCUCUCCGGCCUGACCGCUGGCAUGCACAUGCUAUC ...(((((((((..(((..((..........))..)))..))))))).)).(((..((((((......))))))((((...(((((......)))))...))))..)))... ( -34.80) >DroPse_CAF1 12162 101 - 1 UUCGUCG--CUCUUUU------UCC---UACAGGGGAAUUUGGUAAAUACCAAUUCUUGCAGUUCUUCCUGCAAGUCCUCUCCGCUUUGACCGCAGGCAUGCAUAUGCUCUC ...(((.--.......------...---...((((((..(((((....)))))..(((((((......)))))))))))))..((.......)).)))..((....)).... ( -24.90) >DroEre_CAF1 3838 106 - 1 UAUGACUCUCUAUCUU------ACCCAUUCCAGGGGAGUUUGGAAAGUACCAGUUCCUGCAGUUCUUCCUGCAGGUGCUUUCCGGCCUGACCGCUGGCAUGCACAUGCUAUC ...((((((((.....------..........)))))))).((((((((((......(((((......)))))))))))))))((.....))..((((((....)))))).. ( -34.06) >DroWil_CAF1 3699 96 - 1 UUC----------CUU------GAUCAUUUUAGGGGAAUUUGGUAAAUACCAAUUCCUUCAAUUCUUCCUGCAAGUAUUAUCCGCCUUGACUGCUGGUCUGCAUAUGCUCUC ...----------(((------(......((..(((((((.(((....))))))))))..)).........))))........((...(((.....))).)).......... ( -17.16) >DroYak_CAF1 3804 92 - 1 U--------------U------UCCAAUUCCAGGGGAGUUUGGAAAGUACCAGUUCCUGCAGUUCUUCCUGCAGGUGCUUUCCGGCCUGACCGCUGGCAUGCACAUGCUAUC .--------------.------..........((..((..(((((((((((......(((((......))))))))))))))))..))..))..((((((....)))))).. ( -33.20) >DroPer_CAF1 12128 101 - 1 UUCGUCG--CUCUUUU------UCC---UACAGGGGAAUUUGGUAAAUACCAAUUCUUGCAGUUCUUCCUGCAAGUCCUCUCCGCUUUGACCGCAGGCAUGCAUAUGCUCUC ...(((.--.......------...---...((((((..(((((....)))))..(((((((......)))))))))))))..((.......)).)))..((....)).... ( -24.90) >consensus UUCGUC___CU_UUUU______UCC_AUUACAGGGGAAUUUGGAAAAUACCAAUUCCUGCAGUUCUUCCUGCAAGUGCUCUCCGCCCUGACCGCUGGCAUGCACAUGCUAUC ................................(((((((((((......))))..(((((((......))))))).)))))))............(((((....)))))... (-20.99 = -21.35 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:36 2006