| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,725,767 – 1,725,878 |
| Length | 111 |
| Max. P | 0.703178 |

| Location | 1,725,767 – 1,725,857 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 95.37 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -18.61 |
| Energy contribution | -19.43 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1725767 90 + 27905053 UCAGUUUCUCUUCACUUACGCCGCCGACUGCCGUUGGCCUUUAUUGCCAGCCAUUUUGAAUACCAUUUGGUGGCUUAUUACUUCUGCGGC ...................(((((.((..(((((((((.......))))))..........(((....))))))........)).))))) ( -23.10) >DroSec_CAF1 34892 90 + 1 UCAGAUUCUCUUCACUUACGCCGCCGACUGCCGUUGGCCUUUAUUGCCAGCCAUUUUGAAUACCAUUUGGUGGCUUAUUACUUCUGCGGC ...................(((((.((..(((((((((.......))))))..........(((....))))))........)).))))) ( -23.10) >DroSim_CAF1 36015 90 + 1 UCAGAUUCUCUUCACUUACGCCGCCGACUGCCGUUGGCCUUUAUUGCCAGCCAUUUUGAAUACCAUUUGGUGGCUUAUUACUUCUGCGGC ...................(((((.((..(((((((((.......))))))..........(((....))))))........)).))))) ( -23.10) >DroYak_CAF1 34840 90 + 1 UCAGUUGCUCUUCACUUACGCUACCACUUGCCGUUGGCCUUUAUUGCCAGCCAUUUUGAAUACCAUUUUGUGGCUUAUUACUUCUGCGGC ...((.((...........)).)).....(((((((((.......))))(((((..((.....))....)))))...........))))) ( -17.90) >consensus UCAGAUUCUCUUCACUUACGCCGCCGACUGCCGUUGGCCUUUAUUGCCAGCCAUUUUGAAUACCAUUUGGUGGCUUAUUACUUCUGCGGC ...................(((((.((..(((((((((.......))))))..........(((....))))))........)).))))) (-18.61 = -19.43 + 0.81)

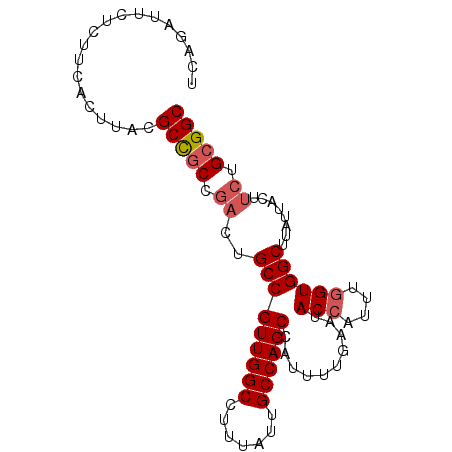
| Location | 1,725,767 – 1,725,857 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 95.37 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -19.66 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
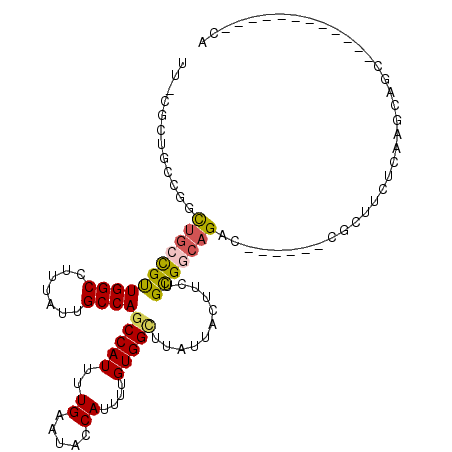
>3R_DroMel_CAF1 1725767 90 - 27905053 GCCGCAGAAGUAAUAAGCCACCAAAUGGUAUUCAAAAUGGCUGGCAAUAAAGGCCAACGGCAGUCGGCGGCGUAAGUGAAGAGAAACUGA (((((.(((....)..((((((....)))........(((((.........)))))..)))..)).)))))...(((........))).. ( -24.30) >DroSec_CAF1 34892 90 - 1 GCCGCAGAAGUAAUAAGCCACCAAAUGGUAUUCAAAAUGGCUGGCAAUAAAGGCCAACGGCAGUCGGCGGCGUAAGUGAAGAGAAUCUGA (((((.(((....)..((((((....)))........(((((.........)))))..)))..)).)))))................... ( -23.70) >DroSim_CAF1 36015 90 - 1 GCCGCAGAAGUAAUAAGCCACCAAAUGGUAUUCAAAAUGGCUGGCAAUAAAGGCCAACGGCAGUCGGCGGCGUAAGUGAAGAGAAUCUGA (((((.(((....)..((((((....)))........(((((.........)))))..)))..)).)))))................... ( -23.70) >DroYak_CAF1 34840 90 - 1 GCCGCAGAAGUAAUAAGCCACAAAAUGGUAUUCAAAAUGGCUGGCAAUAAAGGCCAACGGCAAGUGGUAGCGUAAGUGAAGAGCAACUGA ((((............((((.....))))........(((((.........))))).))))..((....((....)).....))...... ( -19.30) >consensus GCCGCAGAAGUAAUAAGCCACCAAAUGGUAUUCAAAAUGGCUGGCAAUAAAGGCCAACGGCAGUCGGCGGCGUAAGUGAAGAGAAACUGA (((((...(....)..((((((....)))........(((((.........)))))..))).....)))))................... (-19.66 = -19.98 + 0.31)

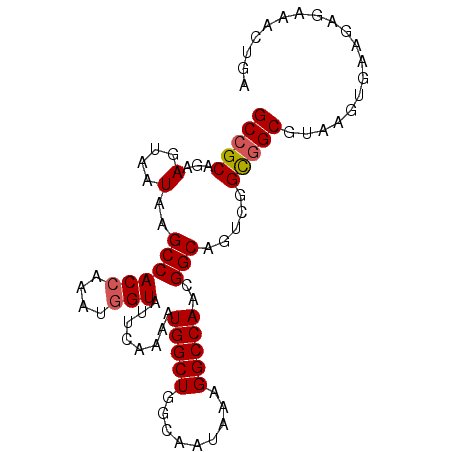
| Location | 1,725,782 – 1,725,878 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.47 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -16.19 |
| Energy contribution | -16.22 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
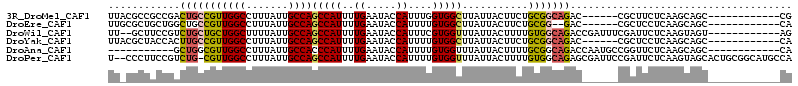
>3R_DroMel_CAF1 1725782 96 + 27905053 UUACGCCGCCGACUGCCGUUGGCCUUUAUUGCCAGCCAUUUUGAAUACCAUUUGGUGGCUUAUUACUUCUGCGGCAGAC------CGCUUCUCAAGCAGC------------CG .......((...(((((((((((.......))))((((((.((.....))...))))))...........)))))))..------.(((.....))).))------------.. ( -27.30) >DroEre_CAF1 34416 94 + 1 UUGCGCUGCUGGCUGCCGUUGGCCUUUAUUGCCAGCCAUUUUGAAUACCAUUUUGUGGCUUAUUACUUCUGCGG--GAC------CGCUCCUCAAGCAGC------------CA ..........((((((.((((((.......))))))....((((...((((...))))............(((.--...------)))...)))))))))------------). ( -27.80) >DroWil_CAF1 30495 100 + 1 UU--GCUUCCGUCUGCUGCUGGCUUUUAUUGCCAGCCAUUUUGAAUACCAUUUCGUGGUUUAUUACUUUUGUGGCAGACCGAUUUCGAUUCUCAAGUAGU------------AG ((--((((..(((((((((((((.......))))))..........(((((...))))).............)))))))((....))......)))))).------------.. ( -27.80) >DroYak_CAF1 34855 96 + 1 UUACGCUACCACUUGCCGUUGGCCUUUAUUGCCAGCCAUUUUGAAUACCAUUUUGUGGCUUAUUACUUCUGCGGCAGAC------CGCUCCUCAAGCAGC------------CA ....(((.....(((((((((((.......))))(((((..((.....))....)))))...........)))))))..------.(((.....))))))------------.. ( -25.30) >DroAna_CAF1 31090 91 + 1 -----------GCUGGCGUUGGCCUUUAUUGCCACCCAUUUUGAAUACCAUUUUGUGGUUUAUUACUUUUGCGGCAGACCAAUGCCGGUUCUCAAGCAGC------------CA -----------(((((((((((......(((((.(...........(((((...)))))...........).))))).)))))))))))...........------------.. ( -25.25) >DroPer_CAF1 30428 111 + 1 U--CCCUUCCGUCUG-CGUUGGCCUUUAUUGCCAGCCAUUUUGAAUACCAUUUUGUGGUUUAUUACUUUUGUGGCAGAGCGAUUCCGAUUCUCAAGUAGCACUGCGGCAUGCCA .--.....((((.((-(..((((.......))))(((((....((((((((...)))))..)))......)))))((((((....)).))))......)))..))))....... ( -23.60) >consensus UU_CGCUGCCGGCUGCCGUUGGCCUUUAUUGCCAGCCAUUUUGAAUACCAUUUUGUGGCUUAUUACUUCUGCGGCAGAC______CGCUUCUCAAGCAGC____________CA ............(((((((((((.......))))(((((..((.....))....)))))...........)))))))..................................... (-16.19 = -16.22 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:33 2006