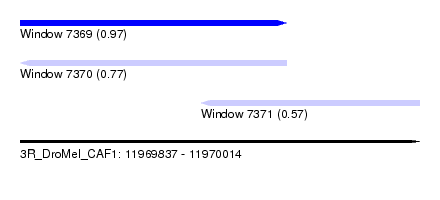
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,969,837 – 11,970,014 |
| Length | 177 |
| Max. P | 0.965995 |

| Location | 11,969,837 – 11,969,955 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 73.23 |
| Mean single sequence MFE | -40.98 |
| Consensus MFE | -33.80 |
| Energy contribution | -34.47 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11969837 118 + 27905053 UAAUUGGCAUGAACGCCACGGUCACCGGCUGGGGUCGUCUCAGCGAGGGCGGCACCCUGCCCUCCGUCCUCCAAGAGGUGAGUGUUAACACGACUUUAAGCUUCAUUUUUAGAAUUCA .....(((......)))(((.(((((.((((((.....))))))((((((((....))))))))............))))).)))................................. ( -38.40) >DroVir_CAF1 85122 96 + 1 UGAUUGGCAUGAAUGCCACGGUCACCGGCUGGGGUCGCCUCAGCGAGGGCGGUACGCUGCCCUCGGUGCUGCAGGAGGUAAGUGG-------AGUU--AGCACAA------------- ....(((((....)))))(((.((((.(((((((...)))))))((((((((....)))))))))))))))..........(((.-------....--..)))..------------- ( -43.60) >DroPse_CAF1 80411 93 + 1 UAAUUGGCAUGAAUGCCACGGUCACCGGCUGGGGUCGUCUGAGCGAGGGCGGCACCCUGCCCUCGGUCCUCCAAGAGGUGAGUGCUUG---------G----------------UGGU ....(((((....)))))((.(((((...(((((......((.(((((((((....))))))))).)))))))...))))).))....---------.----------------.... ( -40.40) >DroGri_CAF1 82817 101 + 1 UGAUUGGCAUGAAUGCCACCGUCACCGGCUGGGGUCGUCUCAGUGAGGGCGGUACGCUGCCGUCGGUGCUGCAGGAGGUAAGUGCA--UGCGACUG--GUCUCAA------------- .(((..(((((..((((.((((((((.((((((.....))))))((.(((((....))))).))))))..)).)).))))....))--))...)..--)))....------------- ( -38.90) >DroAna_CAF1 94924 100 + 1 UAAUUGGCAUGAACGCCACGGUCACCGGUUGGGGUCGCCUCAGCGAGGGCGGCACCCUGCCCUCGGUCCUCCAAGAGGUGAGUUCCAA------------------GUACUGGAUUAG ....((((......))))......(((((((((((((((((...((((((((....))))))))((....))..))))))).))))).------------------..)))))..... ( -44.20) >DroPer_CAF1 79780 93 + 1 UAAUUGGCAUGAAUGCCACGGUCACCGGCUGGGGUCGUCUGAGCGAGGGCGGCACCCUGCCCUCGGUCCUCCAAGAGGUGAGUGCUUG---------G----------------UGGU ....(((((....)))))((.(((((...(((((......((.(((((((((....))))))))).)))))))...))))).))....---------.----------------.... ( -40.40) >consensus UAAUUGGCAUGAAUGCCACGGUCACCGGCUGGGGUCGUCUCAGCGAGGGCGGCACCCUGCCCUCGGUCCUCCAAGAGGUGAGUGCU____________________________U___ ....((((......))))((.(((((.(((((((...)))))))((((((((....))))))))............))))).)).................................. (-33.80 = -34.47 + 0.67)



| Location | 11,969,837 – 11,969,955 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.23 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.63 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11969837 118 - 27905053 UGAAUUCUAAAAAUGAAGCUUAAAGUCGUGUUAACACUCACCUCUUGGAGGACGGAGGGCAGGGUGCCGCCCUCGCUGAGACGACCCCAGCCGGUGACCGUGGCGUUCAUGCCAAUUA ..................(((...((((((....)))...((....))..))).)))(((((..((((((..((((((.(.(.......))))))))..))))))..).))))..... ( -36.00) >DroVir_CAF1 85122 96 - 1 -------------UUGUGCU--AACU-------CCACUUACCUCCUGCAGCACCGAGGGCAGCGUACCGCCCUCGCUGAGGCGACCCCAGCCGGUGACCGUGGCAUUCAUGCCAAUCA -------------(((.((.--....-------(((((((((.....((((...((((((........)))))))))).(((.......))))))))..)))).......)))))... ( -33.00) >DroPse_CAF1 80411 93 - 1 ACCA----------------C---------CAAGCACUCACCUCUUGGAGGACCGAGGGCAGGGUGCCGCCCUCGCUCAGACGACCCCAGCCGGUGACCGUGGCAUUCAUGCCAAUUA ....----------------.---------.......(((((..((((.((..(((((((.((...))))))))).((....))))))))..)))))...(((((....))))).... ( -32.40) >DroGri_CAF1 82817 101 - 1 -------------UUGAGAC--CAGUCGCA--UGCACUUACCUCCUGCAGCACCGACGGCAGCGUACCGCCCUCACUGAGACGACCCCAGCCGGUGACGGUGGCAUUCAUGCCAAUCA -------------.(((...--..((((..--((((.........))))....))))((((..((.(((((.((((((.(.(.......)))))))).))))).))...))))..))) ( -31.40) >DroAna_CAF1 94924 100 - 1 CUAAUCCAGUAC------------------UUGGAACUCACCUCUUGGAGGACCGAGGGCAGGGUGCCGCCCUCGCUGAGGCGACCCCAACCGGUGACCGUGGCGUUCAUGCCAAUUA ...........(------------------((((..(((........)))..)))))(((((..((((((..((((((.((.....))...))))))..))))))..).))))..... ( -38.20) >DroPer_CAF1 79780 93 - 1 ACCA----------------C---------CAAGCACUCACCUCUUGGAGGACCGAGGGCAGGGUGCCGCCCUCGCUCAGACGACCCCAGCCGGUGACCGUGGCAUUCAUGCCAAUUA ....----------------.---------.......(((((..((((.((..(((((((.((...))))))))).((....))))))))..)))))...(((((....))))).... ( -32.40) >consensus ___A____________________________AGCACUCACCUCUUGGAGGACCGAGGGCAGGGUGCCGCCCUCGCUGAGACGACCCCAGCCGGUGACCGUGGCAUUCAUGCCAAUUA .....................................(((((............((((((........))))))((((.(.....).)))).)))))...(((((....))))).... (-25.24 = -25.63 + 0.39)


| Location | 11,969,917 – 11,970,014 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11969917 97 - 27905053 CCAUUUAGAACCACACCGACAAUUUUUUGCGUUCGAUUCGCAGCGGUUAUCAUUGUCGCUGAAUUCUAAAAAUGAAGCUUAAAGUCGUGUUAACACU ...(((((((...((.((((((((..(((((.......)))))..).....))))))).))..)))))))................(((....))). ( -18.70) >DroSec_CAF1 74194 97 - 1 CCAUUUAGAACCACACCGACAAUCUUUUGCGUUCGAUUCGCAGCGGUUAUCAUUGUCGCUGCAUUCUAAAAAUGAAGCUUAAAGUCGUGUUAACACU ...(((((((...((.(((((((((.(((((.......))))).)).....))))))).))..)))))))................(((....))). ( -20.30) >DroSim_CAF1 74521 97 - 1 CCAUUUAGAACCACACCGACAAUCUUUUGCGUUCGAUUCGCAGCGGUUAUCAUUGUCGCUGCAUUCUAAAAAUGAACCUUAAAGUCGUGUUAACACU ...(((((((...((.(((((((((.(((((.......))))).)).....))))))).))..)))))))................(((....))). ( -20.30) >DroEre_CAF1 74935 97 - 1 CCAUUAAGACCCACACCGACAAUCUGUUGCGUUCGAUUCGCAGCGGUUAUCAUUGUCGCUGGAUUCUAAAAAUGGAGCCUAAAGCUGUGUUAACACU (((((.(((.(((...(((((((((((((((.......)))))))).....))))))).)))..)))...)))))(((.....)))(((....))). ( -27.90) >DroYak_CAF1 77611 97 - 1 CCAAUAAGAACCACGCCGACAAUCUGUUGCGUUCGAUUCGCAGCGGUUAUCAUUGUCGCUGGAUUCUAAAAAUGGGCCUUAAAGCCGUGUUAACACU ......(((((((.((.((((((((((((((.......)))))))).....))))))))))).)))).......(((......)))(((....))). ( -30.40) >consensus CCAUUUAGAACCACACCGACAAUCUUUUGCGUUCGAUUCGCAGCGGUUAUCAUUGUCGCUGCAUUCUAAAAAUGAAGCUUAAAGUCGUGUUAACACU ...(((((((...((.(((((((((.(((((.......))))).)).....))))))).))..)))))))................(((....))). (-18.14 = -18.62 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:08 2006