
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,898,535 – 11,898,724 |
| Length | 189 |
| Max. P | 0.985051 |

| Location | 11,898,535 – 11,898,655 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.11 |
| Mean single sequence MFE | -34.01 |
| Consensus MFE | -30.58 |
| Energy contribution | -31.22 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11898535 120 + 27905053 UGCGCUGCUUCUUCUCGAGCAAAUUGUUGCGCGCAUUUUGCUCCUCGAGCAGCAUGCGCAGAUCGUUCAUCUCGUUGGUCAUCUUCUGUGCCUUGCGCUUCCAUUGGCCCACAACCUGAC (((((((((.(((...(((((((.(((.....))).)))))))...))).)))).))))).............((.(((((......((((...))))......))))).))........ ( -38.90) >DroGri_CAF1 4006 120 + 1 UGCGCUGCUUCUUCUCUAGCAAAUUGUUGCGUGCAUUUUGCUCCUCGAGCAGCAUGCGCAAGUCAUUCAUCUCGUUGGUCAUCUUUUGUGCCUUGCGUUUCCAUUGGCCCACAACCUGAC (((((((((.(((....((((((.(((.....))).))))))....))).)))).))))).((((.......(((.(((((.....)).)))..))).......))))............ ( -33.04) >DroEre_CAF1 3066 120 + 1 UGCGCUGCUUCUUCUCGAGCAAAUUGUUGCGCGCAUUUUGCUCCUCGAUUAACAUGCGCAGAUCGUUCAUCUCGUUGGUCAUCUUCUGUGCCUUGCGCUUCCAUUGGCCCACAACCUGAC (((((((.((..((..(((((((.(((.....))).)))))))...))..)))).))))).............((.(((((......((((...))))......))))).))........ ( -31.80) >DroWil_CAF1 3480 120 + 1 UGCGCUGCUUCUUCUCCAGCAAAUUGUUACGGGCAUUUUGCUCUUCGAGCAGCAUGCGCAAAUCAUUCAUCUCAUUGGUCAUCUUUUGUGCCUUGCGCUUCCAUUGGCCCACAACUUGAC (((((((((.(((....((((((.(((.....))).))))))....))).)))).)))))......(((.......(((((......((((...))))......))))).......))). ( -34.74) >DroMoj_CAF1 3417 120 + 1 UGCGCUGCUUCUUCUCCAGCAAAUUGUUUCGUGCAUUUUGCUCCUCGAGCAGCAUGCGCAAAUCAUUCAUCUCGUUGGUCAUCUUUUGAGCCUUGCGUUUCCAUUGGCCCACAACCUGAC (((((((((.(((....((((((.(((.....))).))))))....))).)))).))))).............((.(((((........((...))........))))).))........ ( -32.39) >DroPer_CAF1 3834 120 + 1 UGCGCUGCUUCUUCUCGAGCAAAUUGUUACGGGCAUUCUGCUCCUCGAGCAGCAUGCGCAGAUCGUUCAUCUCGUUUGUCAUCUUCUGUGCCUUGCGCUUCCAUUGGCCCACAACCUGGC .(((((((((......(((((...(((.....)))...)))))...)))))))..(((((((.....((.......))......)))))))...))(((......)))(((.....))). ( -33.20) >consensus UGCGCUGCUUCUUCUCGAGCAAAUUGUUGCGCGCAUUUUGCUCCUCGAGCAGCAUGCGCAAAUCAUUCAUCUCGUUGGUCAUCUUCUGUGCCUUGCGCUUCCAUUGGCCCACAACCUGAC (((((((((.(((....((((((.(((.....))).))))))....))).)))).)))))......(((....((.(((((......((((...))))......))))).))....))). (-30.58 = -31.22 + 0.64)



| Location | 11,898,535 – 11,898,655 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.11 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -30.66 |
| Energy contribution | -30.72 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11898535 120 - 27905053 GUCAGGUUGUGGGCCAAUGGAAGCGCAAGGCACAGAAGAUGACCAACGAGAUGAACGAUCUGCGCAUGCUGCUCGAGGAGCAAAAUGCGCGCAACAAUUUGCUCGAGAAGAAGCAGCGCA ((((..((.((.(((..((....))...))).)).))..)))).....((((.....))))((((.((((.(((((...((.....))..((((....)))))))))....)))))))). ( -37.50) >DroGri_CAF1 4006 120 - 1 GUCAGGUUGUGGGCCAAUGGAAACGCAAGGCACAAAAGAUGACCAACGAGAUGAAUGACUUGCGCAUGCUGCUCGAGGAGCAAAAUGCACGCAACAAUUUGCUAGAGAAGAAGCAGCGCA .(((..(((((((((..((....))...))).((.....)).)).))))..)))......(((((.((((.(((....((((((.((.......)).)))))).)))....))))))))) ( -35.70) >DroEre_CAF1 3066 120 - 1 GUCAGGUUGUGGGCCAAUGGAAGCGCAAGGCACAGAAGAUGACCAACGAGAUGAACGAUCUGCGCAUGUUAAUCGAGGAGCAAAAUGCGCGCAACAAUUUGCUCGAGAAGAAGCAGCGCA ((((..((.((.(((..((....))...))).)).))..)))).....((((.....))))((((.((((..((...(((((((.((.......)).)))))))..))...)))))))). ( -32.70) >DroWil_CAF1 3480 120 - 1 GUCAAGUUGUGGGCCAAUGGAAGCGCAAGGCACAAAAGAUGACCAAUGAGAUGAAUGAUUUGCGCAUGCUGCUCGAAGAGCAAAAUGCCCGUAACAAUUUGCUGGAGAAGAAGCAGCGCA .....(((((((((........(((((((.((.......................)).)))))))....(((((...)))))....))))))))).....((((.........))))... ( -29.70) >DroMoj_CAF1 3417 120 - 1 GUCAGGUUGUGGGCCAAUGGAAACGCAAGGCUCAAAAGAUGACCAACGAGAUGAAUGAUUUGCGCAUGCUGCUCGAGGAGCAAAAUGCACGAAACAAUUUGCUGGAGAAGAAGCAGCGCA ((((..((.((((((..((....))...)))))).))..)))).................(((((.((((.(((....((((((.((.......)).)))))).)))....))))))))) ( -37.10) >DroPer_CAF1 3834 120 - 1 GCCAGGUUGUGGGCCAAUGGAAGCGCAAGGCACAGAAGAUGACAAACGAGAUGAACGAUCUGCGCAUGCUGCUCGAGGAGCAGAAUGCCCGUAACAAUUUGCUCGAGAAGAAGCAGCGCA ((((((((((..(((..((....))...))).((.....)).............)))))))).))..(((((((...(((((((.((.......)).))))))).....).))))))... ( -34.50) >consensus GUCAGGUUGUGGGCCAAUGGAAGCGCAAGGCACAAAAGAUGACCAACGAGAUGAACGAUCUGCGCAUGCUGCUCGAGGAGCAAAAUGCACGCAACAAUUUGCUCGAGAAGAAGCAGCGCA ((((..((.((.(((..((....))...))).)).))..)))).................(((((.((((.(((....((((((.((.......)).)))))).)))....))))))))) (-30.66 = -30.72 + 0.06)

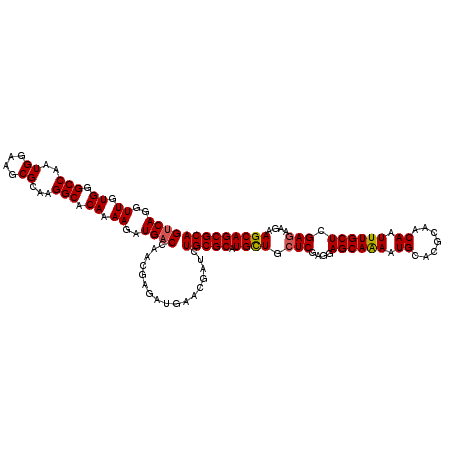

| Location | 11,898,615 – 11,898,724 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.82 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -22.14 |
| Energy contribution | -22.42 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11898615 109 - 27905053 -ACGGAAGUGUAAUAUG-----UUCCACCCCGCCAUUUCAGCUUUCCGAUGCCUACGAAGAAGUUGUGGAACAACGUCAGGUUGUGGGCCAAUGGAAGCGCAAGGCACAGAAGAU -..((((..........-----)))).....(((......((((((....((((((((.((.((((.....)))).))...))))))))....))))))....)))......... ( -30.70) >DroVir_CAF1 3868 105 - 1 -----UAACUUGAUCUGAGCUUUCCC-----ACAUUCGCAGCUAUCCGAUGCCUACGAGGAAGUUGAAGAACAACGUCAGGUUGUGGGCCAAUGGAAACGCAAGGCACAAAAGAU -----...((((.....((((.....-----........)))).((((..((((((((.((.((((.....)))).))...))))))))...))))....))))........... ( -24.22) >DroPse_CAF1 3929 98 - 1 ------AUUGUAAUCCC-----UUUCU------GCUUGUAGCUUUCGGAUGCCUACGAGGAAGUUGAGGAGCAACGCCAGGUUGUGGGCCAAUGGAAGCGCAAGGCACAGAAGAU ------...........-----(((((------((((((.(((((((...((((((((((..((((.....)))).))...))))))))...))))))))))))...)))))).. ( -33.00) >DroWil_CAF1 3560 98 - 1 ------AUUUUACAAUG-----UUU------UCUUUUGCAGCUUUCCGAUGCCUACGAGGAAGUUGAAGAACAACGUCAAGUUGUGGGCCAAUGGAAGCGCAAGGCACAAAAGAU ------...........-----...------(((((((..((((((....((((((((.((.((((.....)))).))...))))))))....))))))((...)).))))))). ( -28.80) >DroMoj_CAF1 3497 111 - 1 AGA--UAAUACAAACAUAACUUUUCCAUG--ACACUUUCAGCUUUCCGAUGCCUACGAAGAAGUUGAGGAACAACGUCAGGUUGUGGGCCAAUGGAAACGCAAGGCUCAAAAGAU ...--..............(((((((.((--(.....)))((((((((..((((((((.((.((((.....)))).))...))))))))...)))))).))..))...))))).. ( -26.90) >DroAna_CAF1 20336 106 - 1 --UAGACCCUUAAUAAU-----UUCUAUCUUUGAA--ACAGCUUUCCGAUGCCUACGAAGAAGUUGAGGAGCAACGUCAAGUUGUGGGCCAAUGGAAGCGCAAGGCUCAGAAGAU --..((.((((.....(-----(((.......)))--)..((((((((..((((((((.((.((((.....)))).))...))))))))...)))))).)))))).))....... ( -29.80) >consensus ______AAUUUAAUAUG_____UUCCA____ACAAUUGCAGCUUUCCGAUGCCUACGAAGAAGUUGAGGAACAACGUCAGGUUGUGGGCCAAUGGAAGCGCAAGGCACAAAAGAU ........................................((((((((..((((((((.((.((((.....)))).))...))))))))...)))))).)).............. (-22.14 = -22.42 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:44 2006