
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,865,944 – 11,866,050 |
| Length | 106 |
| Max. P | 0.834361 |

| Location | 11,865,944 – 11,866,050 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.74 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -23.81 |
| Energy contribution | -23.92 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11865944 106 + 27905053 GCCAUGGCUGGCGGAGCGGCCAGUGAACUGGCUGGCUACCAUCACCAGCACAACGUCAUCCAGGCAGCCAAACUGAUGGCCACCUCGUAAAGUUAAAAAUC-------UGGAC .(((..(((((.(((((((((((....)))))).))).))....)))))..(((.....(.(((..((((......))))..))).)....))).......-------))).. ( -36.90) >DroPse_CAF1 35345 102 + 1 GCCAUGGCGGGGGGAGCGGCCAGUGAGUUGGCCGGAUAUCACCAUCAGCACAAUGUCAUACAGGCGGCCAAGCUGAUGGCCACCUCCUAAGGGCCAUCCCUU----------- ((....)).((((((..((((((.(((.(((((((......))((((((....((((.....)))).....))))))))))).)))))...)))).))))))----------- ( -44.40) >DroWil_CAF1 23893 93 + 1 GCCAUGGCAGGCGGAGGAGCCAGCGAUUUGGCCGGCUAUCAUCAUCAGCAUAAUGUCAUCCAGGCAGCCAAACUAAUGGCGUCUUCUUGAA--------------------AC (((......)))((((((((((....((((((..(((.........)))....((((.....))))))))))....)))).))))))....--------------------.. ( -29.20) >DroYak_CAF1 23414 113 + 1 GCCAUGGCAGGCGGAGCGGCCAGUGAACUGGCAGGCUACCAUCACCAGCACAACGUCAUCCAGGCAGCCAAGCUGAUGGCCACCUCGUAAGGCUAGAACUUUGGGCUCCGGAC (((......)))(((((.(((((....)))))..))).))..................(((.((.((((.......(((((.........)))))........))))))))). ( -40.06) >DroAna_CAF1 23328 101 + 1 GCCAUGGCGGGCGGAGCUGCCAGCGAACUGGCCGGCUACCAUCACCAGCACAACGUCAUCCAGGCGGCCAAGUUGAUGGCCACCUCGUAAGG-----AAUC-------GCGGA ((..(((.((..(((((((((((....)))).))))).)).)).)))))....(((..((((((.(((((......))))).))).....))-----)...-------))).. ( -38.20) >DroPer_CAF1 35448 102 + 1 GCCAUGGCGGGGGGAGCGGCCAGCGAGUUGGCCGGAUAUCACCAUCAGCACAAUGUCAUCCAGGCGGCCAAGCUGAUGGCCACCUCCUAAGGGCCAUCCCUU----------- ((....))((((((..(((((((....))))))).......((((((((....((((.....)))).....))))))))...))))))(((((....)))))----------- ( -45.10) >consensus GCCAUGGCGGGCGGAGCGGCCAGCGAACUGGCCGGCUACCAUCACCAGCACAACGUCAUCCAGGCAGCCAAGCUGAUGGCCACCUCGUAAGG___A_ACUU__________AC ((....))..(((..(.(((((......(((((((......))...........(((.....))))))))......))))).)..)))......................... (-23.81 = -23.92 + 0.11)

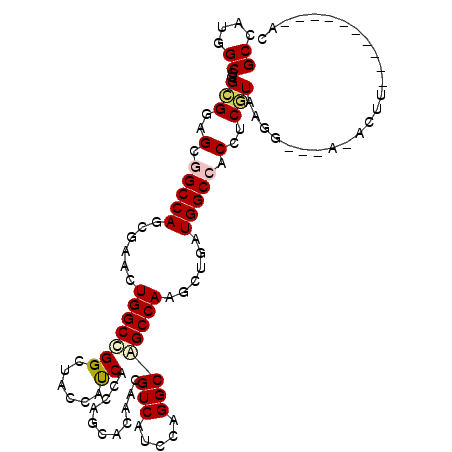

| Location | 11,865,944 – 11,866,050 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.74 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -27.09 |
| Energy contribution | -26.53 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11865944 106 - 27905053 GUCCA-------GAUUUUUAACUUUACGAGGUGGCCAUCAGUUUGGCUGCCUGGAUGACGUUGUGCUGGUGAUGGUAGCCAGCCAGUUCACUGGCCGCUCCGCCAGCCAUGGC ..(((-------......((((.((((.(((..((((......))))..))).).))).)))).(((((((..((..(((((........)))))..)).)))))))..))). ( -44.20) >DroPse_CAF1 35345 102 - 1 -----------AAGGGAUGGCCCUUAGGAGGUGGCCAUCAGCUUGGCCGCCUGUAUGACAUUGUGCUGAUGGUGAUAUCCGGCCAACUCACUGGCCGCUCCCCCCGCCAUGGC -----------(((((....))))).(.(((((((((......))))))))).)..........(((.((((((.....((((((......)))))).......))))))))) ( -42.50) >DroWil_CAF1 23893 93 - 1 GU--------------------UUCAAGAAGACGCCAUUAGUUUGGCUGCCUGGAUGACAUUAUGCUGAUGAUGAUAGCCGGCCAAAUCGCUGGCUCCUCCGCCUGCCAUGGC ((--------------------((.....))))(((((..(((((((((.(((.....((((((....)))))).))).))))))))).((.(((......))).)).))))) ( -28.90) >DroYak_CAF1 23414 113 - 1 GUCCGGAGCCCAAAGUUCUAGCCUUACGAGGUGGCCAUCAGCUUGGCUGCCUGGAUGACGUUGUGCUGGUGAUGGUAGCCUGCCAGUUCACUGGCCGCUCCGCCUGCCAUGGC ....(((((.........((((.((((.(((..((((......))))..))).).))).)))).((..((((((((.....))))..))))..)).)))))(((......))) ( -46.60) >DroAna_CAF1 23328 101 - 1 UCCGC-------GAUU-----CCUUACGAGGUGGCCAUCAACUUGGCCGCCUGGAUGACGUUGUGCUGGUGAUGGUAGCCGGCCAGUUCGCUGGCAGCUCCGCCCGCCAUGGC .((((-------(...-----(.((((.(((((((((......))))))))).).))).)...)))(((((.(((.(((..(((((....))))).))))))..))))).)). ( -45.70) >DroPer_CAF1 35448 102 - 1 -----------AAGGGAUGGCCCUUAGGAGGUGGCCAUCAGCUUGGCCGCCUGGAUGACAUUGUGCUGAUGGUGAUAUCCGGCCAACUCGCUGGCCGCUCCCCCCGCCAUGGC -----------..(((..(((.......(((((((((......)))))))))(((((.(((..(....)..))).)))))(((((......))))))))..))).((....)) ( -45.10) >consensus GU__________AAGU_U___CCUUACGAGGUGGCCAUCAGCUUGGCCGCCUGGAUGACAUUGUGCUGAUGAUGAUAGCCGGCCAAUUCACUGGCCGCUCCGCCCGCCAUGGC ..........................((.((((((((.....(((((((.(((.....(((..(....)..))).))).))))))).....)))))))).))...((....)) (-27.09 = -26.53 + -0.55)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:33 2006