
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,841,145 – 11,841,427 |
| Length | 282 |
| Max. P | 0.955421 |

| Location | 11,841,145 – 11,841,256 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -24.56 |
| Energy contribution | -23.50 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11841145 111 + 27905053 UGGAUUUCCUCAAUUGACGGUGG------UGAUGGUUACACAUAAGCACACACAUUUUUUGGUAUAUUUAUAGCCAUGGUAAAGGUGGAAACCACCUGAAAGCCUCUGAGCUAUAUG .((....))((((..((..((((------(((((......)))...))).))).))..))))......((((((((.(((..((((((...))))))....)))..)).)))))).. ( -26.80) >DroSec_CAF1 21067 115 + 1 UGGAUUUCCCCAAUUGAGGGUGUAUUGAACCAUGGAUACACAUAGGCACACACAUUUUUUGGUAUAUUUAUAGCCAUGGUAAAGGUGGAACCCACCUGA--GCCUCUGAGCUAUAUG .((....))((((..((..((((..((..(((((......))).)))).)))).))..))))......((((((((.(((..((((((...))))))..--)))..)).)))))).. ( -31.00) >DroSim_CAF1 22646 117 + 1 UGGAUUUCCCCAAUUGAGGGUGUAUUGGACCAUGGUUACACAUAGGCACACACAUUUUUUGGUAUAUUUAUAGCCAUGGUAAAGGUGGAACCCACCUGAAAGCCUCUGAGCUAUAUG .((....))((((..((..((((...(..(((((......))).))..))))).))..))))......((((((((.(((..((((((...))))))....)))..)).)))))).. ( -30.90) >DroEre_CAF1 25115 117 + 1 UGGAUUUCCCCAGUUGAGUGUGUAUUGAAAAAUGGCUACACAUGUGUGUGCACAUUAUUUGGUAUAUUUAUAGCCAUGGCAAAGGUGGAACCCACCUGAAAGCCUCCAAGCUAUAUG (((......))).......((((((..((.((((((.((((....)))))).)))).))..)))))).((((((...(((..((((((...))))))....))).....)))))).. ( -36.50) >consensus UGGAUUUCCCCAAUUGAGGGUGUAUUGAACCAUGGUUACACAUAGGCACACACAUUUUUUGGUAUAUUUAUAGCCAUGGUAAAGGUGGAACCCACCUGAAAGCCUCUGAGCUAUAUG .((....))((((..((..((((......(.(((......))).)....)))).))..))))......((((((...(((..((((((...))))))....))).....)))))).. (-24.56 = -23.50 + -1.06)


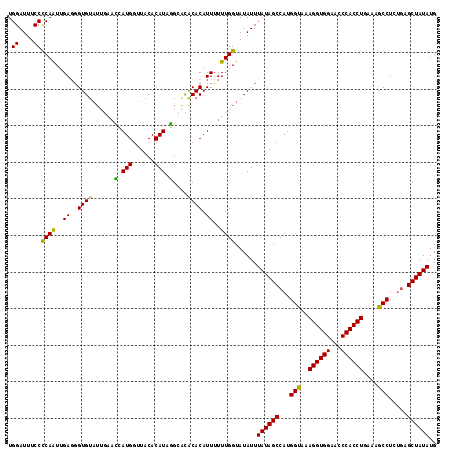
| Location | 11,841,145 – 11,841,256 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -21.88 |
| Energy contribution | -23.00 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11841145 111 - 27905053 CAUAUAGCUCAGAGGCUUUCAGGUGGUUUCCACCUUUACCAUGGCUAUAAAUAUACCAAAAAAUGUGUGUGCUUAUGUGUAACCAUCA------CCACCGUCAAUUGAGGAAAUCCA ..((((((.((..((.....((((((...))))))...)).))))))))...............(((.(((...(((......)))))------))))..........((....)). ( -21.90) >DroSec_CAF1 21067 115 - 1 CAUAUAGCUCAGAGGC--UCAGGUGGGUUCCACCUUUACCAUGGCUAUAAAUAUACCAAAAAAUGUGUGUGCCUAUGUGUAUCCAUGGUUCAAUACACCCUCAAUUGGGGAAAUCCA ..((((((.((..((.--..((((((...))))))...)).))))))))......((((.....((((((..(((((......)))))....))))))......))))((....)). ( -31.50) >DroSim_CAF1 22646 117 - 1 CAUAUAGCUCAGAGGCUUUCAGGUGGGUUCCACCUUUACCAUGGCUAUAAAUAUACCAAAAAAUGUGUGUGCCUAUGUGUAACCAUGGUCCAAUACACCCUCAAUUGGGGAAAUCCA .............((.((((((((((...))))))..(((((((.((((.((((((........)))))).......)))).)))))))........(((......))))))).)). ( -31.40) >DroEre_CAF1 25115 117 - 1 CAUAUAGCUUGGAGGCUUUCAGGUGGGUUCCACCUUUGCCAUGGCUAUAAAUAUACCAAAUAAUGUGCACACACAUGUGUAGCCAUUUUUCAAUACACACUCAACUGGGGAAAUCCA ..(((((((....(((....((((((...))))))..)))..)))))))......(((....(((((....)))))(((((............))))).......)))((....)). ( -31.80) >consensus CAUAUAGCUCAGAGGCUUUCAGGUGGGUUCCACCUUUACCAUGGCUAUAAAUAUACCAAAAAAUGUGUGUGCCUAUGUGUAACCAUGGUUCAAUACACCCUCAAUUGGGGAAAUCCA ..((((((.((..((.....((((((...))))))...)).))))))))......((((.....((((((..(((((......)))))....))))))......))))((....)). (-21.88 = -23.00 + 1.12)

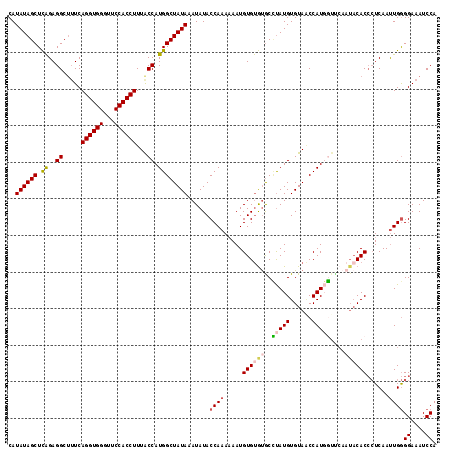
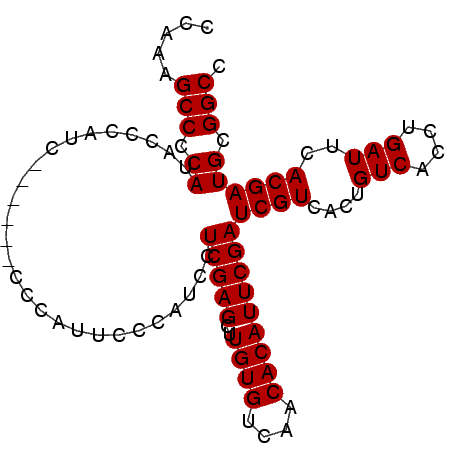
| Location | 11,841,179 – 11,841,294 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.68 |
| Mean single sequence MFE | -32.91 |
| Consensus MFE | -21.69 |
| Energy contribution | -24.08 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11841179 115 + 27905053 CAUAAGCACACACAUUUU---UUGGUAUAUUUAUAGCCAUGGUAAAGGUGGAAACCACCUGAAAGCCUCUGAGCUAUAUGGCACUUGUCAUGAGCGGACUGGCGGUAAAUUUC--AACCU ..................---..(((..((((((.((((.(((..((((((...))))))....))).....(((.((((((....)))))))))....)))).))))))...--.))). ( -32.20) >DroSec_CAF1 21107 113 + 1 CAUAGGCACACACAUUUU---UUGGUAUAUUUAUAGCCAUGGUAAAGGUGGAACCCACCUGA--GCCUCUGAGCUAUAUGGCACUUGUCAUGAGCGGACUGGCGGUAAAUUCC--UGCCU ...(((((..........---..((...((((((.((((.(((..((((((...))))))..--))).....(((.((((((....)))))))))....)))).)))))).))--))))) ( -37.10) >DroSim_CAF1 22686 115 + 1 CAUAGGCACACACAUUUU---UUGGUAUAUUUAUAGCCAUGGUAAAGGUGGAACCCACCUGAAAGCCUCUGAGCUAUAUGGCACUUGUCAUGAGCGGACUGGCGGUAAAUUCC--GGCCU ...((((...........---.(((...((((((.((((.(((..((((((...))))))....))).....(((.((((((....)))))))))....)))).)))))).))--))))) ( -34.80) >DroEre_CAF1 25155 115 + 1 CAUGUGUGUGCACAUUAU---UUGGUAUAUUUAUAGCCAUGGCAAAGGUGGAACCCACCUGAAAGCCUCCAAGCUAUAUGGCACUUGUCAUGAGCGGACUGGCGGUAAAUUCC--GGCCU .(((((....)))))...---..(((..((((((.((((.(((..((((((...))))))....)))(((..(((.((((((....)))))))))))).)))).))))))...--.))). ( -40.30) >DroYak_CAF1 25788 101 + 1 --------------UUAU---UCGGUAUAUUUAUAGCCAUGGUAAAAAUGGAACCCAACUGAAGGCCUCUGAGCUAUAUGGCACUUGUCAUGAGCGGACUGGCGGUAAACUCC--GGCCU --------------...(---(((((..........((((.......))))......))))))((((.....(((.((((((....))))))))).....((.(.....).))--)))). ( -26.39) >DroAna_CAF1 21539 98 + 1 ---A------GGCAUUAUGGACUUGUAUGUUUAUAGUUAUGGUGAA-----ACCCCAUGCGAAAGCCUUCGUCUUAUAUGACACUUGUCAAGAGCUGGCUU--------CUCCGGGGCCG ---.------((((((((((((......))))))))).........-----..(((..(.(((.(((..(.(((....((((....))))))))..)))))--------).).)))))). ( -26.70) >consensus CAUA_GCACACACAUUAU___UUGGUAUAUUUAUAGCCAUGGUAAAGGUGGAACCCACCUGAAAGCCUCUGAGCUAUAUGGCACUUGUCAUGAGCGGACUGGCGGUAAAUUCC__GGCCU .......................(((..((((((.((((.(((..(((((.....)))))....))).....(((.((((((....)))))))))....)))).))))))......))). (-21.69 = -24.08 + 2.39)



| Location | 11,841,294 – 11,841,387 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -15.07 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 1.04 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11841294 93 - 27905053 CCAAAGCCCCAUACCCAUCCCCAUCCCCAUUCCCAUCCUCGAGCUUGUGUCAACACAUUCGAUCGUCACUGUCACCUGAUUCACGAUGCGGCC .....(((.((...........................(((((..((((....)))))))))((((....(((....)))..)))))).))). ( -15.70) >DroSec_CAF1 21220 87 - 1 CCAAAGCCCCAUACCCAUC------CCCAUUCCCAUCCUCGAGCUUGUGUCAACACAUUCGAUCGUCACUGUCACCUGAUUCACGAUGCGGCC .....(((.((........------.............(((((..((((....)))))))))((((....(((....)))..)))))).))). ( -15.70) >DroSim_CAF1 22801 86 - 1 CCAAAGCCCCAUACCCAUC------CCCAUUCCCAUCCUCGAGCUUGUGUCAACACAUUCGAUCGUCACUGUCAC-UGAUUCACGAUGCGGCC .....(((.((........------.............(((((..((((....)))))))))((((....(((..-.)))..)))))).))). ( -13.80) >consensus CCAAAGCCCCAUACCCAUC______CCCAUUCCCAUCCUCGAGCUUGUGUCAACACAUUCGAUCGUCACUGUCACCUGAUUCACGAUGCGGCC .....(((.((...........................(((((..((((....)))))))))((((....(((....)))..)))))).))). (-15.70 = -15.70 + 0.00)


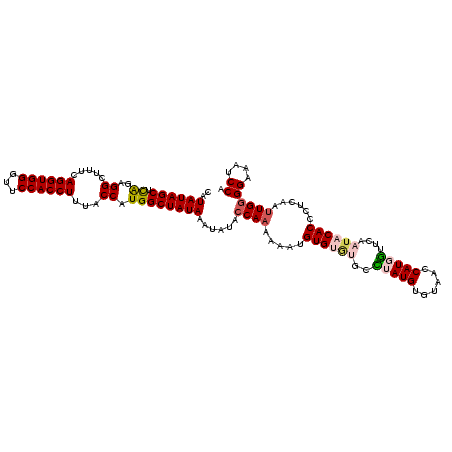
| Location | 11,841,326 – 11,841,427 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.76 |
| Mean single sequence MFE | -20.36 |
| Consensus MFE | -13.72 |
| Energy contribution | -14.05 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11841326 101 - 27905053 AUGACCUCAUUCCGUUGGGGGUGGCACUUCCACUUCCACUCCAAAGCCCCAUACC---------------CAU----CCCCAUCCCCAUUCCCAUCCUCGAGCUUGUGUCAACACAUUCG ...............(((((((((........(((........)))........)---------------)))----)))))................((((..((((....)))))))) ( -19.59) >DroSec_CAF1 21252 95 - 1 AUGACCUCAUUCCGUUGGGGGUGGCACUUCCACUUCCACUCCAAAGCCCCAUACC---------------CAU----C------CCCAUUCCCAUCCUCGAGCUUGUGUCAACACAUUCG ...............(((((((((........(((........)))........)---------------)))----)------))))..........((((..((((....)))))))) ( -19.59) >DroSim_CAF1 22832 95 - 1 AUGACCUCAUUCCGUUGGGGGUGGCACUUCCACUUCCACUCCAAAGCCCCAUACC---------------CAU----C------CCCAUUCCCAUCCUCGAGCUUGUGUCAACACAUUCG ...............(((((((((........(((........)))........)---------------)))----)------))))..........((((..((((....)))))))) ( -19.59) >DroEre_CAF1 25302 103 - 1 AUGACCUCAUUCCGUUGGGGGUGGCACUUCCACUUCCA------------AUUCC-ACUUCCCC--UCCCCAU--ACCCCACACCCCAUUGCCAUCCUCGAAGUUGUGUCAACACAUUCG .((((..((.....((((((((((((............------------.....-........--.......--..............))))))))))))...)).))))......... ( -20.75) >DroYak_CAF1 25921 118 - 1 AUGACCUCAUUCCGUUGGGGGUGGCACUUCCACUUCCUCGCCGUGGCCCCAUACCCCAUUCCCCUUUCCCC--AUACCCCCCUCCCCAGUCCCAUCCUCGAAGUUGUGUCAACACAUUCG .............(..(((((((((..............)))(((....)))...................--..))))))..)..............((((..((((....)))))))) ( -21.14) >DroAna_CAF1 21677 77 - 1 AUGACCUCAUUACGUUGGGGGCGGCACUUCCACC--------AGGGC------------------------AG----C------UCGGGCCCCAUCCUC-AACUUGUGUCAACACAUUUG .((((..((....((((((((.((........))--------.((((------------------------..----.------....))))..)))))-))).)).))))......... ( -21.50) >consensus AUGACCUCAUUCCGUUGGGGGUGGCACUUCCACUUCCACUCCAAAGCCCCAUACC_______________CAU____C______CCCAUUCCCAUCCUCGAACUUGUGUCAACACAUUCG .((((..((.....((((((((((...................................................................))))))))))...)).))))......... (-13.72 = -14.05 + 0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:29 2006