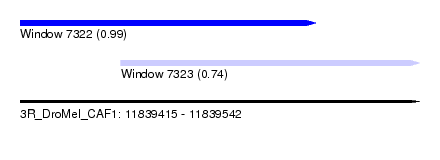
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,839,415 – 11,839,542 |
| Length | 127 |
| Max. P | 0.988022 |

| Location | 11,839,415 – 11,839,509 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.33 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -17.86 |
| Energy contribution | -19.53 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
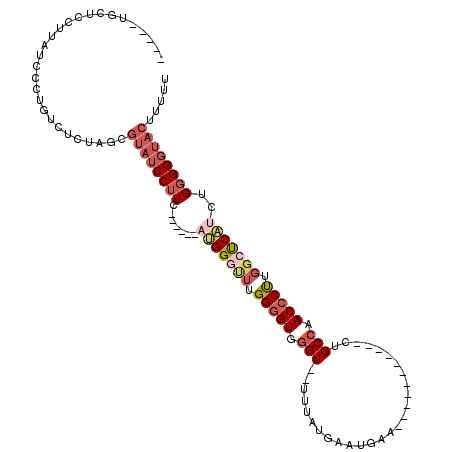
>3R_DroMel_CAF1 11839415 94 + 27905053 -----UGCUCCUUAUCCCUGUCUCUAGCGUAUCCUUC------AUGGGCUUGGGGUUGGU--UUUGUGAAUGAAU------------GCCAACCCUUGGCUCAUCUGGGGGUACUUUUU -----.(((................)))((((((((.------(((((((.(((((((((--(((......))).------------))))))))).)))))))..))))))))..... ( -34.89) >DroSec_CAF1 19324 101 + 1 -----UGCUUCUUAUCCCUGUCUCUA-----UCCAUGAAGGAAAUGGGUUUGGGGUUGGU--UUUGUGAAUGAAUGAC------CCUGCCAACCCUUGGCUCAUCUGGGGGUACUUUUU -----.......(((((((.......-----(((.....))).((((((..(((((((((--...((.(.....).))------...)))))))))..))))))..)))))))...... ( -34.40) >DroSim_CAF1 20814 106 + 1 -----UGCUCCUUAUCCCUGUCUCUAGCGUAUCCUUC------AUGGGCUUGGGGUUGGU--UUUGUGAAUGAAUGAGACGGCGUCUGCCAACCCUUGGCUCAUCUGGGGGUACUUUUU -----.(((................)))((((((((.------(((((((.((((((.((--((..(......)..))))(((....))))))))).)))))))..))))))))..... ( -37.09) >DroEre_CAF1 23318 101 + 1 -----UGGUCCUUAUCCAUGCCCCUAGCGUAUCCUUU------UUGGAUUUGGGGUGGGU--U-UAUGAAUGAA----CCGGCGACUGCCAACCCUUGGCUCAUCUGGGGGUACUUUUU -----(((.......)))(((((((((((.((((...------..)))).))((((.(((--(-((....))))----))(((....))).)))).........)))))))))...... ( -35.50) >DroYak_CAF1 23954 106 + 1 GGUCCUGGUCCUUAUCCCUGUCUCUAGCGUAUCCUUU------UAGGGUUUGGGGUGGGU--U-UAUGAAUGAA----CCGGCCGCUGCCAACCCCUGGCUCAUCUGGGGGUACUUUUU (((..(((((..((((((...((((((.........)------)))))...))))))(((--(-((....))))----)))))))..))).(((((..(.....)..)))))....... ( -39.10) >DroAna_CAF1 19060 80 + 1 -----U-------------GUCUCGAGCGUAUCCUUA------AGGGA--UGGAGUGAGUGCCUUAUGAAUGAA-------------GCCAACCCUUGUCCCUUUGGGGGUUACUUUUC -----.-------------.........(((((((((------(((((--(((..((.((..(........)..-------------))))..))..))))).))))))).)))..... ( -19.70) >consensus _____UGCUCCUUAUCCCUGUCUCUAGCGUAUCCUUC______AUGGGUUUGGGGUGGGU__UUUAUGAAUGAA___________CUGCCAACCCUUGGCUCAUCUGGGGGUACUUUUU ............................((((((((.......(((((((.(((((.(((...........................))).))))).)))))))..))))))))..... (-17.86 = -19.53 + 1.67)


| Location | 11,839,447 – 11,839,542 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -20.98 |
| Energy contribution | -21.81 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11839447 95 + 27905053 ---AUGGGCUUGGGGUUGGU--UUUGUGAAUGAAU------------GCCAACCCUUGGCUCAUCUGGGGGUACUUUUUAUCCUGCCGGAAGUGUC-------CUUUCGACCAAUGGAA ---(((((((.(((((((((--(((......))).------------))))))))).))))))).((((((((.....)))))...((((((....-------)))))).)))...... ( -35.50) >DroSec_CAF1 19354 104 + 1 GAAAUGGGUUUGGGGUUGGU--UUUGUGAAUGAAUGAC------CCUGCCAACCCUUGGCUCAUCUGGGGGUACUUUUUACCCUGCCGGAAGUGUC-------CUUUCGACCAAUGGAA ...((((((..(((((((((--...((.(.....).))------...)))))))))..)))))).((((((((.....)))))...((((((....-------)))))).)))...... ( -37.20) >DroSim_CAF1 20846 107 + 1 ---AUGGGCUUGGGGUUGGU--UUUGUGAAUGAAUGAGACGGCGUCUGCCAACCCUUGGCUCAUCUGGGGGUACUUUUUACCCUGCCGGAAGUGUC-------CUUUCGACCAAUGGAA ---(((((((.((((((.((--((..(......)..))))(((....))))))))).))))))).((((((((.....)))))...((((((....-------)))))).)))...... ( -40.30) >DroEre_CAF1 23350 102 + 1 ---UUGGAUUUGGGGUGGGU--U-UAUGAAUGAA----CCGGCGACUGCCAACCCUUGGCUCAUCUGGGGGUACUUUUUGCCCUGCCGGAAGUCUC-------CGUUCGACCAAUGGAA ---........(((((.(((--(-((....))))----))(((....))).))))).((((..((((((((((.....)))))..)))))))))((-------((((.....)))))). ( -34.80) >DroYak_CAF1 23991 102 + 1 ---UAGGGUUUGGGGUGGGU--U-UAUGAAUGAA----CCGGCCGCUGCCAACCCCUGGCUCAUCUGGGGGUACUUUUUACCCUGCCGGAAGUGUC-------CUUUCGACCAAUGGAA ---..((((..(((((.(((--(-((....))))----))(((....))).)))))..))))...((((((((.....)))))...((((((....-------)))))).)))...... ( -38.70) >DroAna_CAF1 19079 101 + 1 ---AGGGA--UGGAGUGAGUGCCUUAUGAAUGAA-------------GCCAACCCUUGUCCCUUUGGGGGUUACUUUUCACUAGCCCGGAAGUGGCACUCGUACUACUGGCCAAUGAAA ---..((.--(((..((((((((...(((..(((-------------(..((((((((......)))))))).)))))))......(....).))))))))..)))....))....... ( -29.00) >consensus ___AUGGGUUUGGGGUGGGU__UUUAUGAAUGAA___________CUGCCAACCCUUGGCUCAUCUGGGGGUACUUUUUACCCUGCCGGAAGUGUC_______CUUUCGACCAAUGGAA ....((((((.(((((.(((...........................))).))))).))))))..((((((((.....)))))...((((((...........)))))).)))...... (-20.98 = -21.81 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:23 2006