| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,836,299 – 11,836,405 |
| Length | 106 |
| Max. P | 0.737651 |

| Location | 11,836,299 – 11,836,405 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.83 |
| Mean single sequence MFE | -36.95 |
| Consensus MFE | -23.81 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
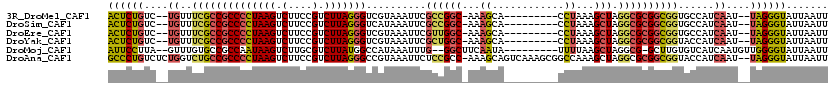
>3R_DroMel_CAF1 11836299 106 + 27905053 ACUCUGUC--UGUUUCGCCGCCCCUAAGUCUUCCGUCUUAGGGUCGUAAAUUCGCCGGC-AAAGCA---------CCUAAAGCUAGGCGCGGCGGUGCCAUCAAU--UAGGGUAUUAAUU ((((((..--((...(((((((((((((.(....).))))))((((.........))))-...((.---------((((....)))).)))))))))....))..--))))))....... ( -37.90) >DroSim_CAF1 17677 106 + 1 ACUCUGUC--UGUUUCGCCGCCCCUAAGUCUUCCGUCUUAGGGUCAUAAAUUCGCCGGC-AAAGCA---------CCUAAAGCUAGGCGCGGCGGUGCCAUCAAU--UAGGGUAUUAAUU ((((((..--((...(((((.(((((((.(....).))))))).)........((((((-..(((.---------......)))..)).))))))))....))..--))))))....... ( -37.50) >DroEre_CAF1 20128 106 + 1 ACUCUGUC--UGUUUCGCCGCCCCUAAGUCUUCCGUCUUAGGGUCGUAAAUUCGUUGGC-AAAGCA---------CCUAAAGCUAGGCGCGGCGGUGCCAUCAAU--UAGGGUAUUAAUU ((((((..--((...(((((((((((((.(....).))))))((((.........))))-...((.---------((((....)))).)))))))))....))..--))))))....... ( -37.00) >DroYak_CAF1 20699 106 + 1 ACUCUGUC--UGUUUCGCCGCCCCUAAGUCUUCCGUCUUAGGGUCGUAAAUUCGCUGGC-AAAGCA---------CCUAAAGCUAGGCGCGGCGGUACCAUCAAU--UAGGGUAUUAAUU ((((((..--((..((((((((((((((.(....).)))))))..........(((...-..))).---------((((....)))).)))))))......))..--))))))....... ( -34.80) >DroMoj_CAF1 19780 106 + 1 AUUCCUUA--GUUUGUGCCGCCAAUAAGUCUUGCGUCUUAUGGCCAUAAAUUUG--GGCUUCAAUA---------UUUUAAGCUAGGCG-GCUUGUGUCAUCAAUGUUGGGGUAUUAAUU ...((..(--(((((((..((((.((((.(....).)))))))))))))))).)--)(((((((((---------((.((((((....)-))))).......)))))))))))....... ( -27.90) >DroAna_CAF1 15443 117 + 1 GCCCUGUCUCUGGUCUGCCGCCCCUAAGUCUUCCGUCUUAGGGCCGUAAAUUCUCCGCC-AAAGCAGUCAAAGCGGCCAAAGCUAGGCGCGGCGGUACCAUCAAU--UAGGGUAUUAAUU ((((((....((((((((((((((((((.(....).)))))((((((.....((.(...-...).)).....)))))).......)).))))))).)))).....--))))))....... ( -46.60) >consensus ACUCUGUC__UGUUUCGCCGCCCCUAAGUCUUCCGUCUUAGGGUCGUAAAUUCGCCGGC_AAAGCA_________CCUAAAGCUAGGCGCGGCGGUGCCAUCAAU__UAGGGUAUUAAUU ((((((....((..((((((((((((((.(....).)))))))..........((((((...((............))...))).))))))))))......))....))))))....... (-23.81 = -24.10 + 0.29)
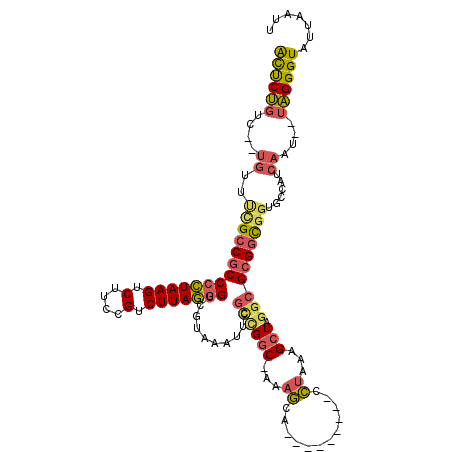
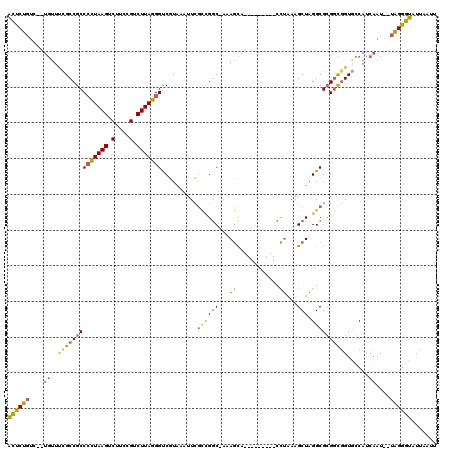
| Location | 11,836,299 – 11,836,405 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.83 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -22.14 |
| Energy contribution | -23.06 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11836299 106 - 27905053 AAUUAAUACCCUA--AUUGAUGGCACCGCCGCGCCUAGCUUUAGG---------UGCUUU-GCCGGCGAAUUUACGACCCUAAGACGGAAGACUUAGGGGCGGCGAAACA--GACAGAGU .((((((......--))))))(((...)))(((((((....))))---------)))(((-((((.((......)).(((((((.(....).))))))).)))))))...--........ ( -36.40) >DroSim_CAF1 17677 106 - 1 AAUUAAUACCCUA--AUUGAUGGCACCGCCGCGCCUAGCUUUAGG---------UGCUUU-GCCGGCGAAUUUAUGACCCUAAGACGGAAGACUUAGGGGCGGCGAAACA--GACAGAGU .((((((......--)))))).....(((((((((.(((......---------.)))..-...)))..........(((((((.(....).))))))))))))).....--........ ( -34.30) >DroEre_CAF1 20128 106 - 1 AAUUAAUACCCUA--AUUGAUGGCACCGCCGCGCCUAGCUUUAGG---------UGCUUU-GCCAACGAAUUUACGACCCUAAGACGGAAGACUUAGGGGCGGCGAAACA--GACAGAGU .((((((......--))))))(((...)))(((((((....))))---------)))(((-(((..((......)).(((((((.(....).)))))))..))))))...--........ ( -34.80) >DroYak_CAF1 20699 106 - 1 AAUUAAUACCCUA--AUUGAUGGUACCGCCGCGCCUAGCUUUAGG---------UGCUUU-GCCAGCGAAUUUACGACCCUAAGACGGAAGACUUAGGGGCGGCGAAACA--GACAGAGU .((((((......--)))))).((..(((((((((((....))))---------)))...-(((..((......))..((((((.(....).)))))))))))))..)).--........ ( -34.30) >DroMoj_CAF1 19780 106 - 1 AAUUAAUACCCCAACAUUGAUGACACAAGC-CGCCUAGCUUAAAA---------UAUUGAAGCC--CAAAUUUAUGGCCAUAAGACGCAAGACUUAUUGGCGGCACAAAC--UAAGGAAU ..........((....(((......)))((-((((..(((..(((---------(.(((.....--)))))))..))).(((((.(....).))))).))))))......--...))... ( -22.20) >DroAna_CAF1 15443 117 - 1 AAUUAAUACCCUA--AUUGAUGGUACCGCCGCGCCUAGCUUUGGCCGCUUUGACUGCUUU-GGCGGAGAAUUUACGGCCCUAAGACGGAAGACUUAGGGGCGGCAGACCAGAGACAGGGC ........((((.--.((..((((.(.(((((((((((.(((..(((((...........-))))).))).))).)))((((((.(....).)))))).))))).)))))..)).)))). ( -44.80) >consensus AAUUAAUACCCUA__AUUGAUGGCACCGCCGCGCCUAGCUUUAGG_________UGCUUU_GCCGGCGAAUUUACGACCCUAAGACGGAAGACUUAGGGGCGGCGAAACA__GACAGAGU .((((((........)))))).....((((((((..(((................)))...))..............(((((((.(....).)))))))))))))............... (-22.14 = -23.06 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:20 2006