
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,814,840 – 11,815,093 |
| Length | 253 |
| Max. P | 0.887900 |

| Location | 11,814,840 – 11,814,959 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.13 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -19.30 |
| Energy contribution | -19.94 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11814840 119 + 27905053 AGCU-CUAGGUACACACGUGGCUAACGACGAAAAGUAAAAACAUAUUUUCGAAAAUUAUAUAACCCAACAAUGGAAGGGACAAUAUUUUCCCUGACGUUGUCAGCGAAAACAAAUUCAUA .(((-...(((.....(((.....))).((((((...........))))))...........)))..((((((..(((((........)))))..)))))).)))(((......)))... ( -25.40) >DroSec_CAF1 19176 120 + 1 AGCGCCUAGGUACAUACGUGGCUAACGACGAAAAGUAAAAACAUAUUUUCGAAAAUUAUAUAACCCAAAAAUGGAAGGGACAAUAUUUUCCCUGACGUUGUCAGCGAAAACAAAUUCAUA ..(((.(((.(((....))).)))..((((((((...........)))))...................((((..(((((........)))))..))))))).))).............. ( -23.50) >DroSim_CAF1 20169 120 + 1 AGCGCCUAGGUACAUACGUGUCUAACGACGAAAAGUAAAAACAUAUUUUCGAAAAUUAUAUAACCCAACAAUGGAAGGGACAAUAUUUUCCCUGACGUUGUCAGCGAAAACAAAUUCAUA ..(((...(((........(((....)))(((((...........)))))............)))..((((((..(((((........)))))..))))))..))).............. ( -26.10) >DroEre_CAF1 19477 115 + 1 AGCG-CUAGGU----ACGUGGCAAAUGACGAAAAGUAAAAACAUAUUUUCCAAAAUUAUAUAACCCAACAAUGGAAGGGACAAUAUUUUCACUGGCGUUGUCAGCGAAAACAAAUUCAUA ((((-((((((----.(((.....)))))(((((...........(((((((...................))))))).......))))).))))))))......(((......)))... ( -18.48) >DroYak_CAF1 19516 115 + 1 AGCG-CUAGGU----ACGUGGCAAACGACGAAAAGUAAAAACAUAUUUUCCAAAAUUAUAUAACCCAACAAUGGAAGGGACAAUAUUUUCCCUGGCGUUGUCAGCGAAAACAAAUUCAUA ..((-((.(((----.(((.....)))..(.((((((......)))))).)...........)))..((((((..(((((........)))))..)))))).)))).............. ( -23.30) >consensus AGCG_CUAGGUACA_ACGUGGCUAACGACGAAAAGUAAAAACAUAUUUUCGAAAAUUAUAUAACCCAACAAUGGAAGGGACAAUAUUUUCCCUGACGUUGUCAGCGAAAACAAAUUCAUA .((.....(((.....(((.....))).((((((...........))))))...........)))..((((((..(((((........)))))..))))))..))(((......)))... (-19.30 = -19.94 + 0.64)

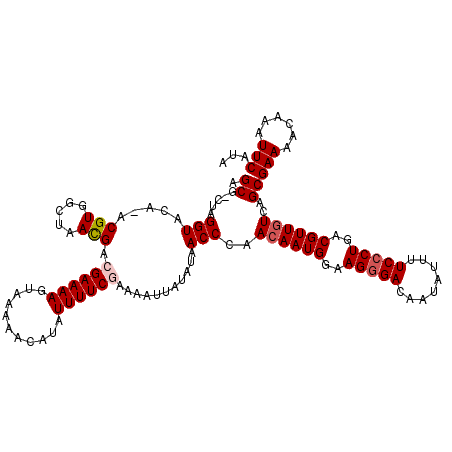
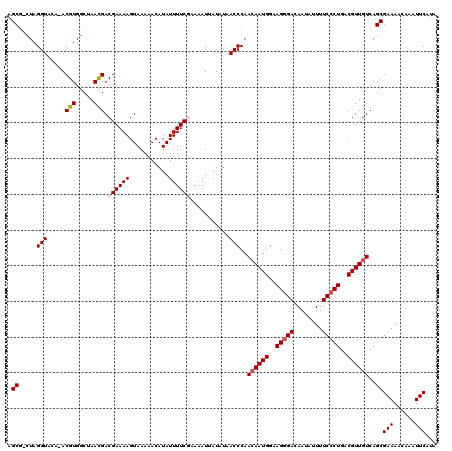
| Location | 11,814,879 – 11,814,999 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -24.38 |
| Energy contribution | -23.82 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11814879 120 + 27905053 ACAUAUUUUCGAAAAUUAUAUAACCCAACAAUGGAAGGGACAAUAUUUUCCCUGACGUUGUCAGCGAAAACAAAUUCAUAAUUCUGUUUAUUUGCUUGGUCCGAGAGCGAAAGGAUCUUG ........................(((((((((..(((((........)))))..)))))).((((((((((............))))..)))))))))..(((((.(....)..))))) ( -26.40) >DroSec_CAF1 19216 120 + 1 ACAUAUUUUCGAAAAUUAUAUAACCCAAAAAUGGAAGGGACAAUAUUUUCCCUGACGUUGUCAGCGAAAACAAAUUCAUAAUUCUGUUUAUUUGCUUGGUCCGAGAGCGAAAGGAUCUCG ........................((((.((((..(((((........)))))..))))....(((((((((............))))..)))))))))..(((((.(....)..))))) ( -25.00) >DroSim_CAF1 20209 120 + 1 ACAUAUUUUCGAAAAUUAUAUAACCCAACAAUGGAAGGGACAAUAUUUUCCCUGACGUUGUCAGCGAAAACAAAUUCAUAAUUCUGUUUAUUUGCUUGGUCCGAGAGCGAAAGGAUCUCG ........................(((((((((..(((((........)))))..)))))).((((((((((............))))..)))))))))..(((((.(....)..))))) ( -28.50) >DroEre_CAF1 19512 120 + 1 ACAUAUUUUCCAAAAUUAUAUAACCCAACAAUGGAAGGGACAAUAUUUUCACUGGCGUUGUCAGCGAAAACAAAUUCAUAAUUCUAUUUAUUUGCUUGGUCCGAGAGCGAAAGGAUCUCG ......((((((...................))))))((((....(((((.(((((...))))).)))))(((((..(((....)))..)))))....))))((((.(....)..)))). ( -26.11) >DroYak_CAF1 19551 120 + 1 ACAUAUUUUCCAAAAUUAUAUAACCCAACAAUGGAAGGGACAAUAUUUUCCCUGGCGUUGUCAGCGAAAACAAAUUCAUAAUUCUGUUUAUUUGCUUGGUCCGAGAGCGAAAGGAUCUCG ........................(((((((((..(((((........)))))..)))))).((((((((((............))))..)))))))))..(((((.(....)..))))) ( -26.90) >consensus ACAUAUUUUCGAAAAUUAUAUAACCCAACAAUGGAAGGGACAAUAUUUUCCCUGACGUUGUCAGCGAAAACAAAUUCAUAAUUCUGUUUAUUUGCUUGGUCCGAGAGCGAAAGGAUCUCG ........................(((....)))...((((....(((((.(((((...))))).)))))(((((..(((....)))..)))))....))))((((.(....)..)))). (-24.38 = -23.82 + -0.56)

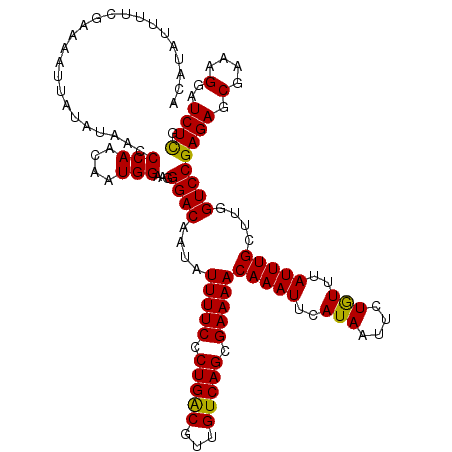
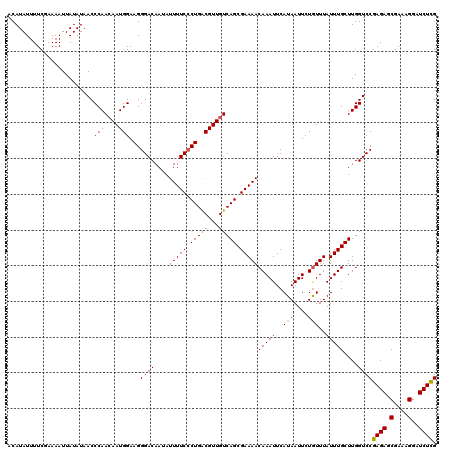
| Location | 11,814,959 – 11,815,053 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -24.59 |
| Energy contribution | -24.67 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11814959 94 + 27905053 AUUCUGUUUAUUUGCUUGGUCCGAGAGCGAAAGGAUCUUGAAGGAC--------------------------AUCGAAGGGAUCGAGGGAACCAAGUGAGCACGAGAUGCGGCAAUAACA .(((((((((((((((((((((((((.(....)..))))((.....--------------------------.))....)))))))).....)))))))))).))).............. ( -24.20) >DroSec_CAF1 19296 94 + 1 AUUCUGUUUAUUUGCUUGGUCCGAGAGCGAAAGGAUCUCGAAGGAC--------------------------AUCGAAGGGACCGAGAGAACCAAGUGAGCACGAGAUGCGGCAAUAACA .(((((((((((((((((((((((((.(....)..))))((.....--------------------------.))....)))))))).....)))))))))).))).............. ( -29.70) >DroSim_CAF1 20289 94 + 1 AUUCUGUUUAUUUGCUUGGUCCGAGAGCGAAAGGAUCUCGAAGGAC--------------------------AUCGAAGGGACCGAGGGAACCAAGUGAGCACGAGAUGCGGCAAUAACA .(((((((((((((((((((((((((.(....)..))))((.....--------------------------.))....)))))))).....)))))))))).))).............. ( -29.70) >DroEre_CAF1 19592 120 + 1 AUUCUAUUUAUUUGCUUGGUCCGAGAGCGAAAGGAUCUCGAAGGACGUUGAGAGGAUAGACGUCGAAGGGACGUCGGAGGGACCGAGGGAACCAAGUGAGCACGAGAUGCGGCAAUAACA ....(((((.(((.((((((((((((.(....)..))))...................((((((.....))))))....)))))))).)))..))))).((.((.....))))....... ( -36.70) >DroYak_CAF1 19631 105 + 1 AUUCUGUUUAUUUGCUUGGUCCGAGAGCGAAAGGAUCUCGAAGGACG---------------UGGAAGGGACGUCGAAGGGUCCGAGGGAACCAAGUGAGCACGAGAUGCGGCAAUAACA .((((((((((((((((((.((((((.(....)..))))....((((---------------(.......)))))....)).))))).....)))))))))).))).............. ( -32.20) >consensus AUUCUGUUUAUUUGCUUGGUCCGAGAGCGAAAGGAUCUCGAAGGAC__________________________AUCGAAGGGACCGAGGGAACCAAGUGAGCACGAGAUGCGGCAAUAACA .(((((((((((((((((((((((((.(....)..))))....((............................))....)))))))).....)))))))))).))).............. (-24.59 = -24.67 + 0.08)

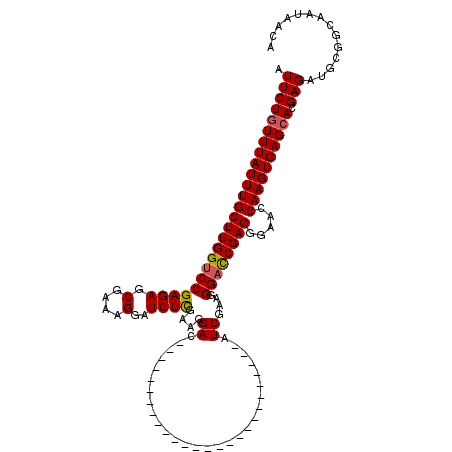

| Location | 11,814,999 – 11,815,093 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -26.02 |
| Energy contribution | -26.26 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11814999 94 + 27905053 AAGGAC--------------------------AUCGAAGGGAUCGAGGGAACCAAGUGAGCACGAGAUGCGGCAAUAACAAUAACAACCGCAUCCCCUGGGUGUCCUGUCUGUUGUCCGC ..((((--------------------------(.((.(((((((.((((......((....))..(((((((...............))))))))))).))).))))...)).))))).. ( -32.66) >DroSec_CAF1 19336 94 + 1 AAGGAC--------------------------AUCGAAGGGACCGAGAGAACCAAGUGAGCACGAGAUGCGGCAAUAACAAUAACAACCGCAUCCCCUGGGUGUCCUGUCCGCUGUCCCC ..((((--------------------------(.((.(((((((.((........((....))..(((((((...............)))))))..)).))).))))...)).))))).. ( -29.36) >DroSim_CAF1 20329 94 + 1 AAGGAC--------------------------AUCGAAGGGACCGAGGGAACCAAGUGAGCACGAGAUGCGGCAAUAACAAUAACAACCGCAUCCCCUGGGUGUCCUGUCCGCUGUCCCC ..((((--------------------------(.((.(((((((.((((......((....))..(((((((...............))))))))))).))).))))...)).))))).. ( -35.96) >DroEre_CAF1 19632 119 + 1 AAGGACGUUGAGAGGAUAGACGUCGAAGGGACGUCGGAGGGACCGAGGGAACCAAGUGAGCACGAGAUGCGGCAAUAACAAUAACAACCGCAUCCCCUGGGUGUCCUGUCCGCUU-CCCC ..(((.((.((.(((((.((((((.....))))))......(((.((((......((....))..(((((((...............))))))))))).)))))))).)).)).)-)).. ( -46.46) >DroYak_CAF1 19671 104 + 1 AAGGACG---------------UGGAAGGGACGUCGAAGGGUCCGAGGGAACCAAGUGAGCACGAGAUGCGGCAAUAACAAUAACAACCGCAUCCCCUGGGUGUCCUGUCCGCUU-CCCC ..(((.(---------------((((.((((((((....)).((.((((......((....))..(((((((...............))))))))))).)))))))).))))).)-)).. ( -40.96) >consensus AAGGAC__________________________AUCGAAGGGACCGAGGGAACCAAGUGAGCACGAGAUGCGGCAAUAACAAUAACAACCGCAUCCCCUGGGUGUCCUGUCCGCUGUCCCC .(((((...........................((....))(((.((((......((....))..(((((((...............))))))))))).))))))))............. (-26.02 = -26.26 + 0.24)

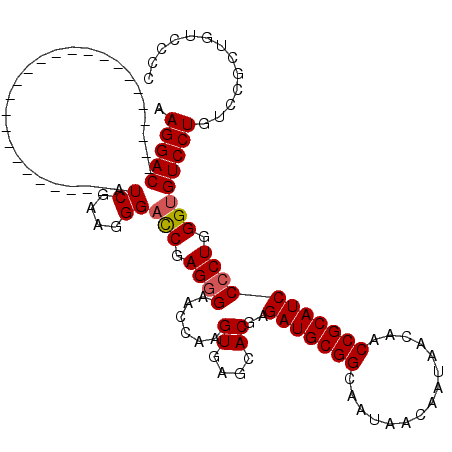
| Location | 11,814,999 – 11,815,093 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -37.96 |
| Consensus MFE | -31.01 |
| Energy contribution | -30.73 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11814999 94 - 27905053 GCGGACAACAGACAGGACACCCAGGGGAUGCGGUUGUUAUUGUUAUUGCCGCAUCUCGUGCUCACUUGGUUCCCUCGAUCCCUUCGAU--------------------------GUCCUU ..(((((.......(((.(((((.((((((((((.((.......)).)))))))))).)).......)))))).((((.....)))))--------------------------)))).. ( -33.61) >DroSec_CAF1 19336 94 - 1 GGGGACAGCGGACAGGACACCCAGGGGAUGCGGUUGUUAUUGUUAUUGCCGCAUCUCGUGCUCACUUGGUUCUCUCGGUCCCUUCGAU--------------------------GUCCUU .((((((.((((..((((...((.((((((((((.((.......)).)))))))))).))(......).........)))).)))).)--------------------------))))). ( -35.50) >DroSim_CAF1 20329 94 - 1 GGGGACAGCGGACAGGACACCCAGGGGAUGCGGUUGUUAUUGUUAUUGCCGCAUCUCGUGCUCACUUGGUUCCCUCGGUCCCUUCGAU--------------------------GUCCUU .((((((.((((..((((....((((((((((((.((.......)).))))))))..((....))......))))..)))).)))).)--------------------------))))). ( -36.30) >DroEre_CAF1 19632 119 - 1 GGGG-AAGCGGACAGGACACCCAGGGGAUGCGGUUGUUAUUGUUAUUGCCGCAUCUCGUGCUCACUUGGUUCCCUCGGUCCCUCCGACGUCCCUUCGACGUCUAUCCUCUCAACGUCCUU ((((-(((.((((.(((.(((((.((((((((((.((.......)).)))))))))).)).......))))))....))))))..((((((.....))))))..)))))........... ( -44.31) >DroYak_CAF1 19671 104 - 1 GGGG-AAGCGGACAGGACACCCAGGGGAUGCGGUUGUUAUUGUUAUUGCCGCAUCUCGUGCUCACUUGGUUCCCUCGGACCCUUCGACGUCCCUUCCA---------------CGUCCUU .(((-(.(.(((..((((...((.((((((((((.((.......)).)))))))))).)).......(((((....))))).......))))..))).---------------).)))). ( -40.10) >consensus GGGGACAGCGGACAGGACACCCAGGGGAUGCGGUUGUUAUUGUUAUUGCCGCAUCUCGUGCUCACUUGGUUCCCUCGGUCCCUUCGAU__________________________GUCCUU .........((((.(((.(((((.((((((((((.((.......)).)))))))))).)).......)))))).((((.....))))...........................)))).. (-31.01 = -30.73 + -0.28)

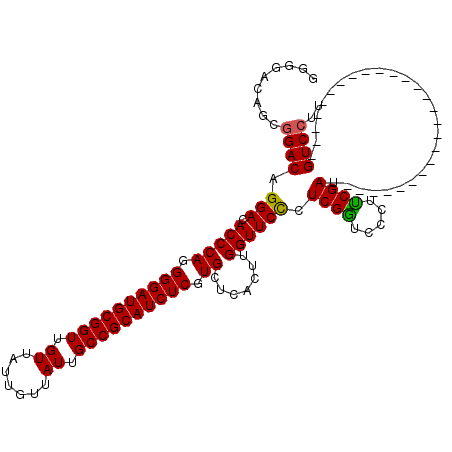

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:07 2006