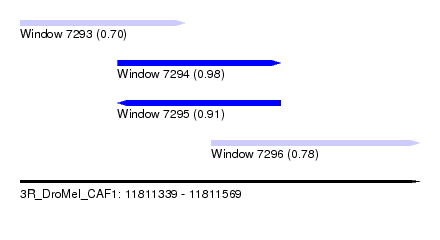
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,811,339 – 11,811,569 |
| Length | 230 |
| Max. P | 0.976459 |

| Location | 11,811,339 – 11,811,434 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -20.44 |
| Energy contribution | -21.68 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11811339 95 + 27905053 --------------AACAGGAACUUAUGUACAUUCCGCUAUAGU--------GGCCAGAUUCCGAAUGCU--GUCGUCACCAAAUC-AAAUCUGGCCAAAGCAGGCCAGAUGAUAAGGAC --------------........(((((...((((..(((....(--------(((((((((.....((.(--(....)).))....-.)))))))))).....)))..)))))))))... ( -24.30) >DroSec_CAF1 15662 117 + 1 AAUACCUCUUUGCGACAAGGAGCUUAUGUACAUCCCGCGAUAGUAGCAAUCUGGUCGGAUUCCCAAUGCU--GUCGUCACCAAAUC-AAAUCUGGCCAAAGCAGGCCAGAUGAUAAGGAC .....(((((((...)))))))..............(((((((((..(((((....))))).....))))--)))))..((..(((-(..(((((((......)))))))))))..)).. ( -35.90) >DroSim_CAF1 16037 117 + 1 AAUACCUCUUUGCGACAAGGAGCUUAUGUACAUGCCGCGAUAGUAGCAAUCUGGUCGGAUUCCCAAUGCU--GUCGUCACCAAAUC-AAAUCUGGCCAAAGCAGGCCAGAUGAUAAGGAC .....(((((((...)))))))..............(((((((((..(((((....))))).....))))--)))))..((..(((-(..(((((((......)))))))))))..)).. ( -35.90) >DroEre_CAF1 15958 114 + 1 AAUACCUUUUUGCGACAAGGAACUU----ACAUCCCUCGAUUGUGGCAAUCUGGUCGGAUUCCCGAUGCU--GUCGUCACCAAAUCAAAAUCUGGCCAAUCCAGGCCAGAUGAUAAGGAC ....((((.((((((((.(((....----...)))....((((.((.(((((....)))))))))))..)--)))))............((((((((......)))))))))).)))).. ( -34.00) >DroYak_CAF1 15944 116 + 1 AAUACCUCUUUGCGCCAAGGAACUU----ACAUGCCGCGAUAGUGGCAAUCUGGUUGGAUUCCCGAGGCUGUGUCGUCACCAAAUCCAAAUCUGGCCAAACCAGGCCAGAUGAUAAGGAC ....(((..((.......(((....----...((((((....))))))...(((((((....))))(((......))).)))..)))..((((((((......))))))))))..))).. ( -37.30) >consensus AAUACCUCUUUGCGACAAGGAACUUAUGUACAUCCCGCGAUAGUAGCAAUCUGGUCGGAUUCCCAAUGCU__GUCGUCACCAAAUC_AAAUCUGGCCAAAGCAGGCCAGAUGAUAAGGAC ....................................(((((...((((....((.......))...))))..)))))..((..(((...((((((((......)))))))))))..)).. (-20.44 = -21.68 + 1.24)

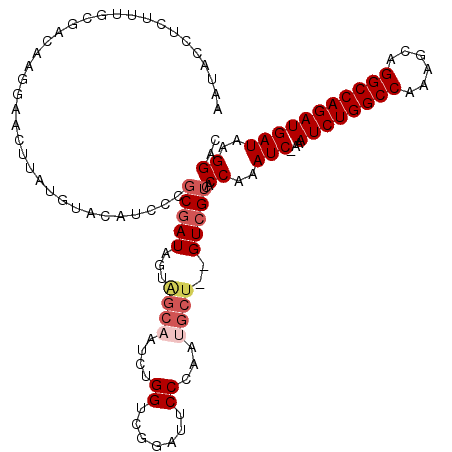
| Location | 11,811,395 – 11,811,489 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.85 |
| Mean single sequence MFE | -40.36 |
| Consensus MFE | -29.60 |
| Energy contribution | -30.20 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11811395 94 + 27905053 CAAAUC-AAAUCUGGCCAAAGCAGGCCAGAUGAUAAGGAC----------------------CUAUGGCA---UAUACUCACCAAGUCGGAGAGCGAGAGUUGCCCCGAUAAGGAUAGUC ...(((-(..(((((((......))))))))))).....(----------------------((.(((..---........))).(((((.((((....))).).))))).)))...... ( -28.50) >DroSec_CAF1 15740 116 + 1 CAAAUC-AAAUCUGGCCAAAGCAGGCCAGAUGAUAAGGACUCGGCUGGGCAUAUGACUAGGGCUAUGGCA---UAUACUCGCCAAGUCGGAGAGCGAGAGUUGCCCCGAUAAGGAUAGUC ...(((-(..(((((((......)))))))))))...((((.(((.(((.(((((.(((......)))))---))).))))))..(((((.((((....))).).)))))......)))) ( -40.70) >DroSim_CAF1 16115 116 + 1 CAAAUC-AAAUCUGGCCAAAGCAGGCCAGAUGAUAAGGACUCGGGUGGGCAUAUGGCUAUGGCUAUGGCA---UAUACUCGCCAAGUCGGAGAGCGAGAGUUGCCCCGAUAAGGAUAGUC ...(((-(..(((((((......)))))))))))...((((((((((.((.((((((....)))))))).---..))))))((..(((((.((((....))).).)))))..))..)))) ( -44.00) >DroEre_CAF1 16032 117 + 1 CAAAUCAAAAUCUGGCCAAUCCAGGCCAGAUGAUAAGGACCCGGGUGAGCAUAUGGCUAUGGCUAUGGCA---UAUACACGCCAAGUCGGAGAGCGAGAGUUGCCCCGAUAAGGAUAGUC ...(((...((((((((......))))))))..........((((..(((...(.(((.(((((.((((.---.......)))))))))...))).)..)))..)))).....))).... ( -40.10) >DroYak_CAF1 16020 120 + 1 CAAAUCCAAAUCUGGCCAAACCAGGCCAGAUGAUAAGGACCCGGGUGGGCAUAUGGCUAUGGCUAUGGCUAUGGCUACACGCCAAGUCGGAGAGCGAGAGUUGCCCCGAUAAGGAUAGUC ...((((..((((((((......)))))))).....((..((((.(.(((...((((((((((....))))))))))...))).).))))..(((....)))...)).....)))).... ( -48.50) >consensus CAAAUC_AAAUCUGGCCAAAGCAGGCCAGAUGAUAAGGACCCGGGUGGGCAUAUGGCUAUGGCUAUGGCA___UAUACUCGCCAAGUCGGAGAGCGAGAGUUGCCCCGAUAAGGAUAGUC .........((((((((......)))))))).............................(((((((((...........)))..(((((.((((....))).).)))))....)))))) (-29.60 = -30.20 + 0.60)

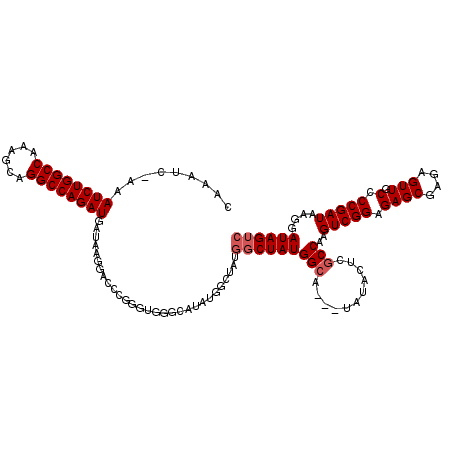
| Location | 11,811,395 – 11,811,489 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.85 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -27.70 |
| Energy contribution | -28.34 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11811395 94 - 27905053 GACUAUCCUUAUCGGGGCAACUCUCGCUCUCCGACUUGGUGAGUAUA---UGCCAUAG----------------------GUCCUUAUCAUCUGGCCUGCUUUGGCCAGAUUU-GAUUUG ((((..((.....))((((...(((((.(........))))))....---))))...)----------------------)))(..(((((((((((......)))))))..)-)))..) ( -31.00) >DroSec_CAF1 15740 116 - 1 GACUAUCCUUAUCGGGGCAACUCUCGCUCUCCGACUUGGCGAGUAUA---UGCCAUAGCCCUAGUCAUAUGCCCAGCCGAGUCCUUAUCAUCUGGCCUGCUUUGGCCAGAUUU-GAUUUG .............(((((.......)))))..((((((((..(((((---((...((....))..)))))))...))))))))(..(((((((((((......)))))))..)-)))..) ( -39.70) >DroSim_CAF1 16115 116 - 1 GACUAUCCUUAUCGGGGCAACUCUCGCUCUCCGACUUGGCGAGUAUA---UGCCAUAGCCAUAGCCAUAUGCCCACCCGAGUCCUUAUCAUCUGGCCUGCUUUGGCCAGAUUU-GAUUUG ...........((((((((...(((((.(........))))))....---)))).....((((....)))).....))))...(..(((((((((((......)))))))..)-)))..) ( -35.40) >DroEre_CAF1 16032 117 - 1 GACUAUCCUUAUCGGGGCAACUCUCGCUCUCCGACUUGGCGUGUAUA---UGCCAUAGCCAUAGCCAUAUGCUCACCCGGGUCCUUAUCAUCUGGCCUGGAUUGGCCAGAUUUUGAUUUG .............(((((.......)))))..(((((((.(((((((---((...((....))..)))))))..))))))))).(((..((((((((......))))))))..))).... ( -35.10) >DroYak_CAF1 16020 120 - 1 GACUAUCCUUAUCGGGGCAACUCUCGCUCUCCGACUUGGCGUGUAGCCAUAGCCAUAGCCAUAGCCAUAUGCCCACCCGGGUCCUUAUCAUCUGGCCUGGUUUGGCCAGAUUUGGAUUUG ...........(((((((.......)).).))))...(((((((.((.((.((....)).)).)).)))))))....(((((((.....((((((((......))))))))..))))))) ( -39.50) >consensus GACUAUCCUUAUCGGGGCAACUCUCGCUCUCCGACUUGGCGAGUAUA___UGCCAUAGCCAUAGCCAUAUGCCCACCCGAGUCCUUAUCAUCUGGCCUGCUUUGGCCAGAUUU_GAUUUG ((((..((.....))((((......((((.((.....)).))))......)))).........................))))......((((((((......))))))))......... (-27.70 = -28.34 + 0.64)


| Location | 11,811,449 – 11,811,569 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -39.91 |
| Energy contribution | -40.15 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11811449 120 + 27905053 ACCAAGUCGGAGAGCGAGAGUUGCCCCGAUAAGGAUAGUCGCGCGACCGACGCGUGAUGCAACUUAAUCAAAUCGUGUUUUGGUAUCGCCCACCCUGCCUCUCGCUCUAUCUGUGCCAGC .....(((((((((((((((((((.((.....))...((((((((.....)))))))))))))...((((((......))))))...............))))))))..)))).)).... ( -39.20) >DroSec_CAF1 15816 120 + 1 GCCAAGUCGGAGAGCGAGAGUUGCCCCGAUAAGGAUAGUCGCGCGACCGACGCGUGAUGCAACUUAAUCAAAUCGUGUUUUGGUAUCACUCUCCCUGCCUCUCGCUCUAUCUCUGCCGAC .....(((((((((((((((((((.((.....))...((((((((.....)))))))))))))...((((((......))))))...............))))))))).......))))) ( -42.91) >DroSim_CAF1 16191 120 + 1 GCCAAGUCGGAGAGCGAGAGUUGCCCCGAUAAGGAUAGUCGCGCGACCGACGCGUGAUGCAACUUAAUCAAAUCGUGUUUUGGUAUCACCCUCCCUGCCUCUCGCUCUAUCUCUGCCGGC (((.((....((((((((((((((.((.....))...((((((((.....)))))))))))))...((((((......))))))...............)))))))))....))...))) ( -42.80) >DroEre_CAF1 16109 120 + 1 GCCAAGUCGGAGAGCGAGAGUUGCCCCGAUAAGGAUAGUCGCGCGACCGACGCGUGAUGCAACUUAAUCAAAUCGUGUUUUGGUAUCGCCCUCCCUGCCUCUCGCUCUAUCUGUGCCGGC (((....(((((((((((((((((.((.....))...((((((((.....)))))))))))))...((((((......))))))...............))))))))..))))....))) ( -43.10) >DroYak_CAF1 16100 120 + 1 GCCAAGUCGGAGAGCGAGAGUUGCCCCGAUAAGGAUAGUCGCGCAACCGACGCGUGAUGAAACUUAAUCAAAUCGUGUUUUGGUAUCGCCCACCCUGCCUCUCGCUCUAUCUGUGCCGGC (((....(((((((((((((..((.....((((....(((((((.......)))))))....))))((((((......))))))............)))))))))))..))))....))) ( -40.00) >consensus GCCAAGUCGGAGAGCGAGAGUUGCCCCGAUAAGGAUAGUCGCGCGACCGACGCGUGAUGCAACUUAAUCAAAUCGUGUUUUGGUAUCGCCCUCCCUGCCUCUCGCUCUAUCUGUGCCGGC .....(((((((((((((((..((.....((((....((((((((.....))))))))....))))((((((......))))))............)))))))))))).......))))) (-39.91 = -40.15 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:56 2006