
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,810,430 – 11,810,655 |
| Length | 225 |
| Max. P | 0.558399 |

| Location | 11,810,430 – 11,810,538 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.12 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -23.02 |
| Energy contribution | -22.94 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11810430 108 - 27905053 AGACAUUGCGAUAAGGAUGCAAGGGUGCGGGUGCGGGUGCUGGACUAUCAGUAGUAG------CAGUA------GCAAAGUCCGGAAGAUGCCUUGUGGUCAGAAGAUAAAAGGCCAAAG .(((.((((........((((....))))(.(((.(.(((((......))))).).)------)).).------)))).)))(....)..(((((...(((....)))..)))))..... ( -29.40) >DroSec_CAF1 14706 107 - 1 AGACCUUGCGAUAAGAAUGCAAGGGUGCGGGUGCGGGUGCUGG-CUAUCAGUAGUUG------CAGUA------GCAAAGUCCGGAAGAUGCCUUGUGGUCAGAAGAUAAAAGGCCAAAG ...(((((((.......))))))).(((.(.(((((.(((((.-....))))).)))------)).).------))).....(....)..(((((...(((....)))..)))))..... ( -35.00) >DroSim_CAF1 15104 107 - 1 AGACGUUGCGAUAAGGAUGCAAGGGUGCGGGUGCGGGUGCUGG-CUAUCAGUAGUUG------CAGUA------GCAAAGUCCGGAAGAUGCCUUGUGGUCAGAAGAUAAAAGGCCAAAG .(((.((((........((((....))))(.(((((.(((((.-....))))).)))------)).).------)))).)))(....)..(((((...(((....)))..)))))..... ( -32.20) >DroEre_CAF1 15059 107 - 1 AGUCAUUGCGAUAAGGAUGCAAGGGUGC------GGGUGCUGG-CUAUCAGCAGUAG------CAGUAGCAGUUGCAAAGUCCGGAAGAUGCCUUGUGGUCAAAAGAUAAAAGGCCAGAG .(((.(((((.......)))))((((((------((.((((((-((((.....))))------)..))))).)))))...)))....)))(((((...(((....)))..)))))..... ( -31.00) >DroYak_CAF1 15028 119 - 1 AGGCAUUGCGAUAAGGAUGCAAGGGUGCGGGUGCGGGUGCUGG-CUAUCAGUAGUAGCAGUGGCAGUAGCAGUUGCAAAGUCCGGAAGAUGCCUUGUGGUCAGAAGAUAAAAGGCCAAAG (((((((.......((((.......(((((.(((.(.((((.(-((((.....)))))...)))).).))).)))))..))))....)))))))..(((((...........)))))... ( -38.80) >consensus AGACAUUGCGAUAAGGAUGCAAGGGUGCGGGUGCGGGUGCUGG_CUAUCAGUAGUAG______CAGUA______GCAAAGUCCGGAAGAUGCCUUGUGGUCAGAAGAUAAAAGGCCAAAG .(((.((((.........(((....))).......(.((((((....)))))).)...................)))).)))(....)..(((((...(((....)))..)))))..... (-23.02 = -22.94 + -0.08)

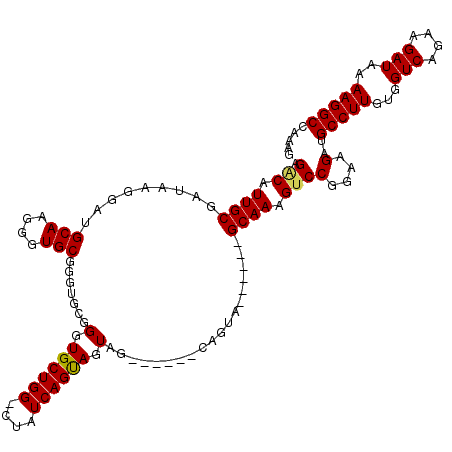

| Location | 11,810,498 – 11,810,615 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.55 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -28.90 |
| Energy contribution | -29.50 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11810498 117 + 27905053 GCACCCGCACCCGCACCCUUGCAUCCUUAUCGCAAUGUCUCCUCCUCGACUCCUCCCCGCAUGGGUGACCCGCACCCUGUGGCCAGGGCCUCCUCC---UCUCCCUACUGGCUGGGUCGG ....(((.(((((((....))).........((...(((........)))((((..(((((.(((((.....))))))))))..))))........---...........)).))))))) ( -35.50) >DroSec_CAF1 14773 117 + 1 GCACCCGCACCCGCACCCUUGCAUUCUUAUCGCAAGGUCUCCUCCUCGACUCCUCCCCGCAUGGGUGACCCGCACCCUGUGGCCAGGGCCUCCUCC---UCUCCCUACUGGCUGGGUCGG ....(((.((((....((((((.........))))))..............(((..(((((.(((((.....))))))))))..)))(((......---..........))).))))))) ( -39.89) >DroSim_CAF1 15171 117 + 1 GCACCCGCACCCGCACCCUUGCAUCCUUAUCGCAACGUCUCCUCCUCGACUCCUCCCCGCAUGGGUGACCCGCAGCCUGUGGCCAGGGCCUCCUCC---UCUCCCUACUGGCUGGGUCGG ....(((.((((.(((((.(((...(.....)....(((........)))........))).)))))......((((.((((..((((.......)---)))..)))).))))))))))) ( -35.70) >DroEre_CAF1 15132 114 + 1 GCACCC------GCACCCUUGCAUCCUUAUCGCAAUGACUCCUCCUCGACUACUCCCCGCAUGGGUGACCCGCACCCUGUGGCCAGGGCCUCCUCCUCUCCUCUCUACUGGUUGGGUCGG ....((------(.((((((((.........))))............(((((....(((((.(((((.....))))))))))..((((.....))))...........)))))))))))) ( -30.60) >DroYak_CAF1 15107 117 + 1 GCACCCGCACCCGCACCCUUGCAUCCUUAUCGCAAUGCCUACUCCUCGGCUCCUCCCCGCAUGGGUGACCCGCACCCUGUGGCCAGGGCCUCCUCC---CCUCCCUACUAGCUGGGUCGG ....(((.((((((....((((.........))))............(((.(((..(((((.(((((.....))))))))))..))))))......---...........)).))))))) ( -39.80) >consensus GCACCCGCACCCGCACCCUUGCAUCCUUAUCGCAAUGUCUCCUCCUCGACUCCUCCCCGCAUGGGUGACCCGCACCCUGUGGCCAGGGCCUCCUCC___UCUCCCUACUGGCUGGGUCGG ..........(((.((((..((..............(((........)))((((..(((((.(((((.....))))))))))..))))......................)).))))))) (-28.90 = -29.50 + 0.60)



| Location | 11,810,538 – 11,810,655 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.52 |
| Mean single sequence MFE | -43.42 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.04 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11810538 117 + 27905053 CCUCCUCGACUCCUCCCCGCAUGGGUGACCCGCACCCUGUGGCCAGGGCCUCCUCC---UCUCCCUACUGGCUGGGUCGGAUCUUGGUCUGCCCGGCGAGGCGCCGACUUAAAGUUUUCC .....(((...(((..(((((.(((((.....))))))))))..)))(((((....---.....(....)((((((.(((((....))))))))))))))))..)))............. ( -46.40) >DroSec_CAF1 14813 112 + 1 CCUCCUCGACUCCUCCCCGCAUGGGUGACCCGCACCCUGUGGCCAGGGCCUCCUCC---UCUCCCUACUGGCUGGGUCGGAUCUUGGUCUG-----CGAGGCGCCGACUUAAAGUUUUCC .....(((.(.((((.(((((.(((((.....))))))))))((((((..(((..(---((..((....))..)))..)))))))))....-----.)))).).)))............. ( -38.80) >DroSim_CAF1 15211 112 + 1 CCUCCUCGACUCCUCCCCGCAUGGGUGACCCGCAGCCUGUGGCCAGGGCCUCCUCC---UCUCCCUACUGGCUGGGUCGGAUCUUGGUCUG-----CGAGGCGCCGACUUAAAGUUUUCC .....(((.(.(((((((((..((....)).))((((.((((..((((.......)---)))..)))).))))))).(((((....)))))-----.)))).).)))............. ( -36.40) >DroEre_CAF1 15166 120 + 1 CCUCCUCGACUACUCCCCGCAUGGGUGACCCGCACCCUGUGGCCAGGGCCUCCUCCUCUCCUCUCUACUGGUUGGGUCGGAUCUCGGUCUGCCCGCCGAGGAGCCGACUUAAAGUUUUCC .((((((.....((..(((((.(((((.....))))))))))..))(((..........((........))..(((.(((((....)))))))))))))))))................. ( -43.20) >DroYak_CAF1 15147 117 + 1 ACUCCUCGGCUCCUCCCCGCAUGGGUGACCCGCACCCUGUGGCCAGGGCCUCCUCC---CCUCCCUACUAGCUGGGUCGGAUCUUGGUCUGCCCGCCGAGGAGCCGACUUAAAGUUUUCC .....((((((((((.(((((.(((((.....))))))))))...((((..((..(---(..(((........)))..)).....))...))))...))))))))))............. ( -52.30) >consensus CCUCCUCGACUCCUCCCCGCAUGGGUGACCCGCACCCUGUGGCCAGGGCCUCCUCC___UCUCCCUACUGGCUGGGUCGGAUCUUGGUCUGCCCG_CGAGGCGCCGACUUAAAGUUUUCC ...(((((........(((((.(((((.....))))))))))...(((((...(((......(((........)))..)))....)))))......)))))................... (-33.00 = -33.04 + 0.04)


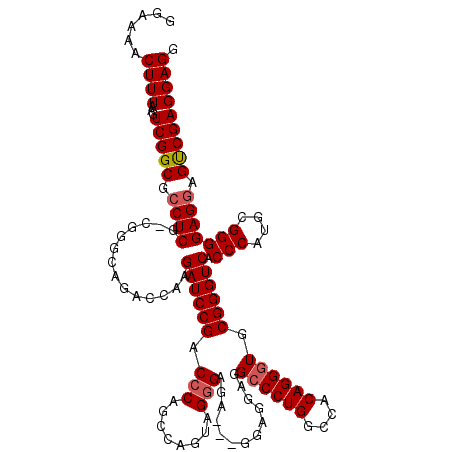
| Location | 11,810,538 – 11,810,655 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.52 |
| Mean single sequence MFE | -47.68 |
| Consensus MFE | -40.48 |
| Energy contribution | -40.92 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11810538 117 - 27905053 GGAAAACUUUAAGUCGGCGCCUCGCCGGGCAGACCAAGAUCCGACCCAGCCAGUAGGGAGA---GGAGGAGGCCCUGGCCACAGGGUGCGGGUCACCCAUGCGGGGAGGAGUCGAGGAGG ......((((...(((((.((((((.(((..((((....(((..(((........)))...---)))....((((((....))))))...)))).)))..))))))....))))))))). ( -47.50) >DroSec_CAF1 14813 112 - 1 GGAAAACUUUAAGUCGGCGCCUCG-----CAGACCAAGAUCCGACCCAGCCAGUAGGGAGA---GGAGGAGGCCCUGGCCACAGGGUGCGGGUCACCCAUGCGGGGAGGAGUCGAGGAGG ......((((...(((((.((((.-----..((((....(((..(((........)))...---)))....((((((....))))))...)))).(((....))))))).))))))))). ( -44.80) >DroSim_CAF1 15211 112 - 1 GGAAAACUUUAAGUCGGCGCCUCG-----CAGACCAAGAUCCGACCCAGCCAGUAGGGAGA---GGAGGAGGCCCUGGCCACAGGCUGCGGGUCACCCAUGCGGGGAGGAGUCGAGGAGG ......((((...(((((.(((((-----((........(((..(((........)))...---)))((.((((((((((...))))).))))).))..)))))))....))))))))). ( -43.20) >DroEre_CAF1 15166 120 - 1 GGAAAACUUUAAGUCGGCUCCUCGGCGGGCAGACCGAGAUCCGACCCAACCAGUAGAGAGGAGAGGAGGAGGCCCUGGCCACAGGGUGCGGGUCACCCAUGCGGGGAGUAGUCGAGGAGG .................(((((((((((.....))..((((((((((..((........)).........(((....)))...)))).)))))).(((....))).....))))))))). ( -47.80) >DroYak_CAF1 15147 117 - 1 GGAAAACUUUAAGUCGGCUCCUCGGCGGGCAGACCAAGAUCCGACCCAGCUAGUAGGGAGG---GGAGGAGGCCCUGGCCACAGGGUGCGGGUCACCCAUGCGGGGAGGAGCCGAGGAGU .....(((((...((((((((((........((((....(((..(((..((....))..))---)..))).((((((....))))))...)))).(((....)))))))))))))))))) ( -55.10) >consensus GGAAAACUUUAAGUCGGCGCCUCG_CGGGCAGACCAAGAUCCGACCCAGCCAGUAGGGAGA___GGAGGAGGCCCUGGCCACAGGGUGCGGGUCACCCAUGCGGGGAGGAGUCGAGGAGG ......((((...(((((.((((..............((((((.(((........))).............((((((....)))))).)))))).(((....))))))).))))))))). (-40.48 = -40.92 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:51 2006