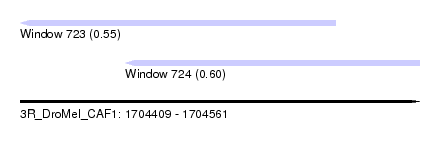
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,704,409 – 1,704,561 |
| Length | 152 |
| Max. P | 0.598610 |

| Location | 1,704,409 – 1,704,529 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
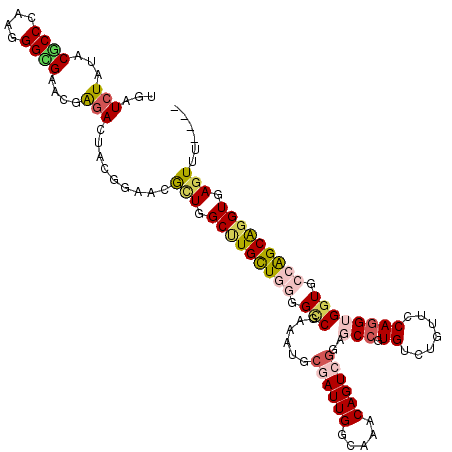
| Mean single sequence MFE | -43.92 |
| Consensus MFE | -29.26 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
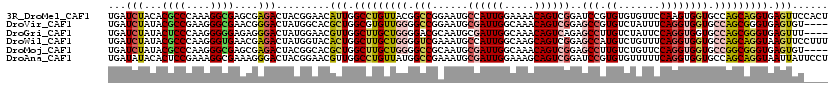
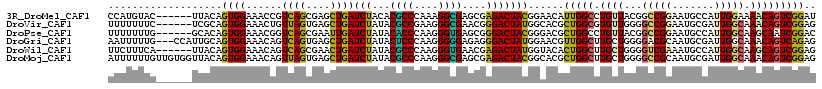
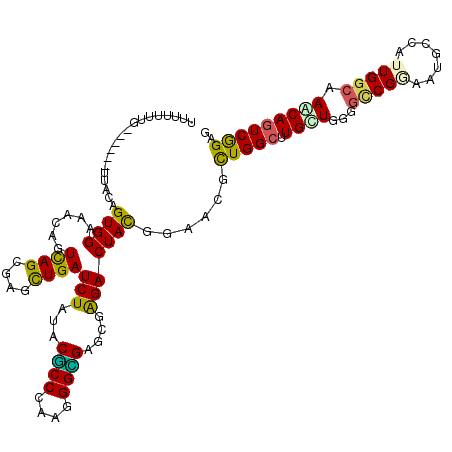
>3R_DroMel_CAF1 1704409 120 - 27905053 UGAUCUACACGCCCAAAGGCGAGCGAGACUACGGAACAUUGGCCUGUUACGGCCGGAAUGCCAUUGGAAAACAGUCGGAUCCGUGUGUGUUCCAAGUGGUGCCAGCAGGUGAGUUCCACU ...(((...((((....))))....)))....(((((..(.(((((((..(((......((((((((((.(((..((....))..))).)))).)))))))))))))))).))))))... ( -42.20) >DroVir_CAF1 17984 116 - 1 UGAUCUAUACGCCGAAGGGCGAACGGGACUAUGGCACGCUGGCGUGUUGGGGCCGGAAUGCGAUUGGCAAACAGUCGGAGCCGUGUCUAUUUCAGGUGGUGCCAGCGGGUGAGUGU---- ...(((...((((....))))....))).......(((((.((.((((((.(((....(.((((((.....)))))).)(((.((.......)))))))).)))))).)).)))))---- ( -45.40) >DroGri_CAF1 17489 116 - 1 UGAUCUAUACUCCCAAGGGGGAGAGGGACUAUGGAACGUUGGCUUGCUGGGGACGCAAUGCGAUUGGCAAACAGUCAGAGCCUUGUCUAUUCCAGGUGGUGCCAGCGGGUGAGUUU---- ...((((((.((((..........)))).)))))).((((((((..((.((((.((((.((..(((((.....))))).)).))))...)))).))..).))))))).........---- ( -40.70) >DroWil_CAF1 11393 120 - 1 UGAUCUAUACGCCCAAGGGUGAACGAGACUAUGGUACACUGGCUUGCUGGGGUCGAAAUGCCAUUGGCAAGCAGUCGGAGCCAUGUCUGUUUCAGGUGGUGCCAGCAGGUAAGUUCCUUU .........((((....))))(((.((((.(((((.(((((.(((((..((((......))).)..))))))))).)..))))))))))))..(((...((((....))))....))).. ( -41.10) >DroMoj_CAF1 17933 116 - 1 UGAUCUAUACGCCCAAGGGCGAGCGAGACUACGGCACGCUGGCUUGCUGGGGCCGCAAUGCGAUUGGCAAACAGUCGGAGCCUUGUCUGUUCCAGGUGGUGCCGGCGGGUGAGUGU---- .........(((((..((((.((((.(.(....)).)))).))))(((((.(((((.....(((((.....)))))(((((.......)))))..))))).)))))))))).....---- ( -53.50) >DroAna_CAF1 11177 120 - 1 UGAUAUACACUCCGAAAGGCGAAAGGGACUACGGAACGUUGGCCUGUUAUGGCCGAAAUGCGAUUGGAAAGCAGUCGGAUCCGUGUGUUUUUCAGGUGGUGCCAGCAGGUAAUUAUUCCU .......((((((....)).((((((.((.(((((.(.((((((......))))))....((((((.....))))))).))))))).)))))).)))).((((....))))......... ( -40.60) >consensus UGAUCUAUACGCCCAAGGGCGAACGAGACUACGGAACGCUGGCUUGCUGGGGCCGAAAUGCGAUUGGCAAACAGUCGGAGCCGUGUCUGUUCCAGGUGGUGCCAGCAGGUGAGUUU____ ...(((...((((....))))....))).........(((.(((((((((.(((......((((((.....))))))..(((.((.......)))))))).))))))))).)))...... (-29.26 = -29.90 + 0.64)
| Location | 1,704,449 – 1,704,561 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -22.84 |
| Energy contribution | -21.85 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1704449 112 - 27905053 CCAUGUAC------UUACAGUGGAAACCGUCAGCGAGCUGAUCUACACGCCCAAAGGCGAGCGAGACUACGGAACAUUGGCCUGUUACGGCCGGAAUGCCAUUGGAAAACAGUCGGAU ((.(((..------...((((((...(((((((....))))(((...((((....))))....)))...)))....((((((......))))))....))))))....)))...)).. ( -33.10) >DroVir_CAF1 18020 112 - 1 UUUUUUUC------UCGCAGUGGAAACUGUUAGUGAGCUGAUCUAUACGCCGAAGGGCGAACGGGACUAUGGCACGCUGGCGUGUUGGGGCCGGAAUGCGAUUGGCAAACAGUCGGAG .......(------(((((((....)))))..((..((..(((....((((....))))..(((..(((..(((((....))))))))..)))......)))..))..)).....))) ( -41.10) >DroPse_CAF1 11416 112 - 1 UUUUUUUG------GCACAGUGGAAACGGUCAGCGAAUUGAUCUAUACACCCAAGGGUGAGCGGGACUACGGGACGCUGGCCUGUUACGGCCGGAAUGCCAUUGGCAAGCAAUCGGAC ...(((((------((...((((....((((((((......(((...((((....))))....)))(....)..))))))))..)))).)))))))((((...))))........... ( -37.60) >DroGri_CAF1 17525 115 - 1 AAUUUUUG---CCAUUGCAGUGGAAACAGUCAGUGAGCUGAUCUAUACUCCCAAGGGGGAGAGGGACUAUGGAACGUUGGCUUGCUGGGGACGCAAUGCGAUUGGCAAACAGUCAGAG ....((((---((((((((.((....)).((((..(((..(((((((.((((..........)))).))))))...)..)))..))))........))))).)))))))......... ( -41.90) >DroWil_CAF1 11433 112 - 1 UUCUUUCA------UUACAGUGGAAACAGUCAGCGAACUGAUCUAUACGCCCAAGGGUGAACGAGACUAUGGUACACUGGCUUGCUGGGGUCGAAAUGCCAUUGGCAAGCAGUCGGAG .(((..(.------...(((((....(((((((....))))).....((((....))))..........))...)))))((((((..((((......))).)..)))))).)..))). ( -34.40) >DroMoj_CAF1 17969 118 - 1 AUUUUUUGUUGUGGUUACAGUGGAAACAGUUAGUGAGCUGAUCUAUACGCCCAAGGGCGAGCGAGACUACGGCACGCUGGCUUGCUGGGGCCGCAAUGCGAUUGGCAAACAGUCGGAG .......((((((((..(((((....)((((((((.((((.(((...((((....))))....)))...)))).)))))))).))))..)))))))).((((((.....))))))... ( -50.30) >consensus UUUUUUUG______UUACAGUGGAAACAGUCAGCGAGCUGAUCUAUACGCCCAAGGGCGAGCGAGACUACGGAACGCUGGCUUGCUGGGGCCGGAAUGCCAUUGGCAAACAGUCGGAG ...................((((......((((....))))(((...((((....))))....)))))))......(((((.((((...(((((.......))))).))))))))).. (-22.84 = -21.85 + -0.99)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:23 2006