| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,781,848 – 11,781,961 |
| Length | 113 |
| Max. P | 0.888978 |

| Location | 11,781,848 – 11,781,961 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.70 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -17.63 |
| Energy contribution | -16.30 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11781848 113 + 27905053 AAGUCAAAGG-CAAAGGCAAAGGCUCAAGCCGGCAAAGAGCUGGGCCAGAAGAUCUCCGAAUCGGAAUUCGUUUCGGAUGACUUCCAGGCCUCGAGCGAAUCGGACUUGGCUGA .((((((..(-(....))....((((....((((.....))))((((.((((.((((((((.((.....)).)))))).))))))..))))..)))).........)))))).. ( -39.70) >DroPse_CAF1 137660 94 + 1 AAAGC--------------------AAGGCCGGCACGGGCAUGAAACAGAAAAUCUCUGAAUCGGAAUUCGUUUCCGACGACUUUCAGACCACAACCGAUUCAGAUUUGGCUGA ..(((--------------------...(((......)))......((((.....((((((((((...(((((...)))))(.....).......))))))))))))))))).. ( -24.60) >DroEre_CAF1 115516 101 + 1 AAGUCAAAGG-------------CACAUGCCAGCAAAGAGCUGGGCCAGAAGGUCUCCGAAUCGGAAUUCGUUUCGGAUGACUUCCAGGCCUCGAGCGAAUCGGACUUGGCUGA .((((((.((-------------(....)))........((((((((.(.(((((((((((.((.....)).)))))).))))).).)))))..))).........)))))).. ( -37.70) >DroYak_CAF1 119150 101 + 1 AAGUCAAAGG-------------CUCAUGCCGGCAAAGAGCUGGGCCAGAAGAUCUCCGAAUCGGAAUUCGUUUCGGAUGACUUCCAGGCCUCGAGCGAAUCGGACUUGGCUGA (((((....(-------------(((....((((.....))))((((.((((.((((((((.((.....)).)))))).))))))..))))..))))......)))))...... ( -37.10) >DroAna_CAF1 118536 107 + 1 AAGCCCACGGGCAAUGGC-------GACGGUGGCAAAGAGAUGUCUCAGAAAAUCUCCGAAUCGGAAUUCGUUUCGGACGAUUUUCAGACCACCUGCGAAUCGGACUUGGCUGA ..(((....)))....((-------(..((((((...(((....))).(((((((((((((.((.....)).)))))).))))))).).))))))))...((((......)))) ( -38.60) >DroPer_CAF1 134786 94 + 1 AAAGC--------------------AAGGCCGGCACGGGCAUGAAACAGAAAAUCUCUGAAUCGGAAUUCGUUUCCGACGACUUUCAGACCACAACCGAUUCAGAUUUGGCUGA ..(((--------------------...(((......)))......((((.....((((((((((...(((((...)))))(.....).......))))))))))))))))).. ( -24.60) >consensus AAGUCAAAGG_______________CAGGCCGGCAAAGAGAUGGGCCAGAAAAUCUCCGAAUCGGAAUUCGUUUCGGACGACUUCCAGACCACGAGCGAAUCGGACUUGGCUGA ..............................((((.(((..........(.....)(((((.((((((.(((((...))))).)))).(........))).)))))))).)))). (-17.63 = -16.30 + -1.33)


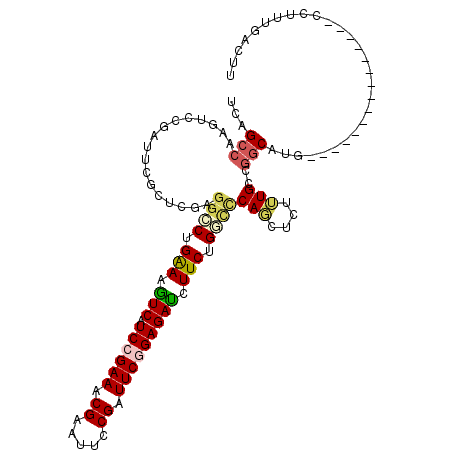
| Location | 11,781,848 – 11,781,961 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.70 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -18.18 |
| Energy contribution | -18.32 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11781848 113 - 27905053 UCAGCCAAGUCCGAUUCGCUCGAGGCCUGGAAGUCAUCCGAAACGAAUUCCGAUUCGGAGAUCUUCUGGCCCAGCUCUUUGCCGGCUUGAGCCUUUGCCUUUG-CCUUUGACUU ...((.(((..(((...((((((((((.(((((((.((((((.((.....)).)))))))).))))))))((.((.....)).)))))))))..))).))).)-)......... ( -38.10) >DroPse_CAF1 137660 94 - 1 UCAGCCAAAUCUGAAUCGGUUGUGGUCUGAAAGUCGUCGGAAACGAAUUCCGAUUCAGAGAUUUUCUGUUUCAUGCCCGUGCCGGCCUU--------------------GCUUU ...(((...((((((((((((((..(((((......))))).))))...))))))))))............(((....)))..)))...--------------------..... ( -26.90) >DroEre_CAF1 115516 101 - 1 UCAGCCAAGUCCGAUUCGCUCGAGGCCUGGAAGUCAUCCGAAACGAAUUCCGAUUCGGAGACCUUCUGGCCCAGCUCUUUGCUGGCAUGUG-------------CCUUUGACUU ......(((((.((..(((....((((.(((((((.((((((.((.....)).)))))))).)))))))))((((.....))))....)))-------------..)).))))) ( -35.90) >DroYak_CAF1 119150 101 - 1 UCAGCCAAGUCCGAUUCGCUCGAGGCCUGGAAGUCAUCCGAAACGAAUUCCGAUUCGGAGAUCUUCUGGCCCAGCUCUUUGCCGGCAUGAG-------------CCUUUGACUU ......(((((......(((.(.((((.(((((((.((((((.((.....)).)))))))).)))))))))))))........(((....)-------------))...))))) ( -35.30) >DroAna_CAF1 118536 107 - 1 UCAGCCAAGUCCGAUUCGCAGGUGGUCUGAAAAUCGUCCGAAACGAAUUCCGAUUCGGAGAUUUUCUGAGACAUCUCUUUGCCACCGUC-------GCCAUUGCCCGUGGGCUU ..((((..(..((((..((.((((((..(((((((.((((((.((.....)).))))))))))))).(((....)))...))))))...-------)).))))..)...)))). ( -39.00) >DroPer_CAF1 134786 94 - 1 UCAGCCAAAUCUGAAUCGGUUGUGGUCUGAAAGUCGUCGGAAACGAAUUCCGAUUCAGAGAUUUUCUGUUUCAUGCCCGUGCCGGCCUU--------------------GCUUU ...(((...((((((((((((((..(((((......))))).))))...))))))))))............(((....)))..)))...--------------------..... ( -26.90) >consensus UCAGCCAAGUCCGAUUCGCUCGAGGCCUGAAAGUCAUCCGAAACGAAUUCCGAUUCGGAGAUCUUCUGGCCCAGCUCUUUGCCGGCAUG_______________CCUUUGACUU ...(((.................((((.(((.(((.((((((.((.....)).))))))))).))).))))(((....)))..)))............................ (-18.18 = -18.32 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:39 2006