| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,780,651 – 11,780,766 |
| Length | 115 |
| Max. P | 0.929780 |

| Location | 11,780,651 – 11,780,766 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.19 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.65 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11780651 115 - 27905053 ---G--GCAACACUAGCAUUGUACUUACAUCUUGGGCACCAGCUCGAAUCGCAUCGUCUCGAGCGCCUUGUCUUCGGCCAUAAUGGCUAUGGCCGUAGGAUAGGCGGUGUCGUAGCUAAA ---(--....)..((((..(((....))).....((((((.((((((..((...))..))))))((((.(((((((((((((.....)))))))).)))))))))))))))...)))).. ( -50.90) >DroVir_CAF1 143239 102 - 1 ---G--GCU-------------ACUCACAUCUUGGGCACCAGCUCAAAGCGCAUCGUUUCGAGCGCCUUAUCCUCGGCCAUAAUGGCUAUGGCAGUGGGAUAUGCGAUGUCAUAACUAAA ---.--...-------------....(((((((((((....)))))).((((.(((...)))))))..((((((..((((((.....))))))...))))))...))))).......... ( -32.80) >DroGri_CAF1 145287 110 - 1 ---GUUGUUUA-------GUCCACUCACAUCUUGGGCACCAAUUCGAAGCGCAUCGUCUCGAGCGCCUUAUCCUCGGCCAUAAUGGCUAUGGCAGUGGGAUAUGCGGUGUCAUAGUUAAA ---........-------................((((((..(((((..((...))..))))).((..((((((..((((((.....))))))...)))))).))))))))......... ( -34.40) >DroWil_CAF1 92010 103 - 1 ---G--A------------UGUACUCACAUUUUGGGCACCAAUUCGAAACGCAUUGUUUCUAGCGCCUUGUCCUCGGCCAUAAUAGCUAUGGCUGUUGGGUAAGCAGAAUCAUAACUAAA ---(--(------------(((....)))))((((......(((((((((.....)))))....((.((..((.((((((((.....))))))))..))..)))).)))).....)))). ( -26.40) >DroMoj_CAF1 153585 108 - 1 ---G--GCCGC-------GCGCACUUACAUCUUGGGCACCAACUCGAAGCGCAUCGUUUCGAGCGCUUUGUCCUCGGCCAUAAUGGCUAUGGCAGUGGGAUAUGCAGUGUCAUAGCUAAA ---(--((((.-------((.((.........)).)).......((((((((.(((...)))))))))))....)))))....(((((((((((.((.......)).))))))))))).. ( -43.80) >DroAna_CAF1 117318 117 - 1 AUUU--AAAACCCUUUC-UGAUACUUACAUCUUGGGCACCAGCUCGAAUCGCAUCGUCUCCAGCGCCUUGUCCUCAGCCAUAAUGGCUAUGGCCGUGGGAUAAGCGGUAUCGUAGCUAAA ....--....(((....-.(((......)))..)))....((((((((((((..(((.....)))..((((((((.((((((.....)))))).).)))))))))))).))).))))... ( -33.10) >consensus ___G__GCA_C________UGUACUCACAUCUUGGGCACCAACUCGAAGCGCAUCGUCUCGAGCGCCUUGUCCUCGGCCAUAAUGGCUAUGGCAGUGGGAUAUGCGGUGUCAUAGCUAAA ..................................((((((..(((((.(((...))).))))).((..((((((..((((((.....))))))...)))))).))))))))......... (-22.70 = -23.65 + 0.95)

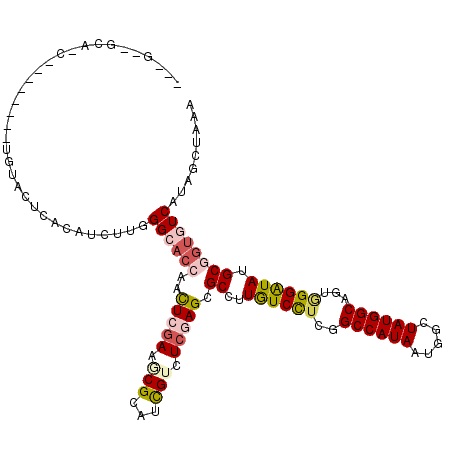

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:37 2006