
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,713,520 – 11,713,650 |
| Length | 130 |
| Max. P | 0.940410 |

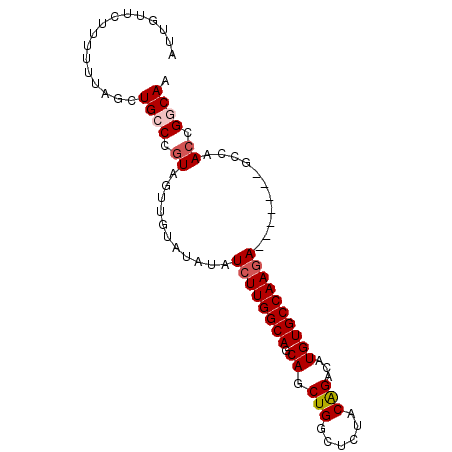
| Location | 11,713,520 – 11,713,610 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -19.35 |
| Energy contribution | -19.91 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11713520 90 + 27905053 AUUGUUCUUUUUUAGCUGCCCGUAGUUGUAUAUAUCUUGGCAGCAGCUGGCUCUACAGACAUGUGCCAACAGCCAAGAGCCAACCGGCAA .....((((.....((((((.(((........)))...)))))).((((....((((....))))....)))).))))(((....))).. ( -24.20) >DroSec_CAF1 50809 90 + 1 AUUGUUCUUUUUUAGCUGCCCGUGGUUGUAUAUAUCUUGGCAGCAGCUGGCUCUACAGACAUGUGCCAAGAGCCAAGAGCCAAGCGGCAA ..............(((((...(((((.(.....((((((((.((.(((......)))...))))))))))....).))))).))))).. ( -32.40) >DroSim_CAF1 52483 83 + 1 AUUGUUCUUUUUUAGCUGCCCGUGGUUGUAUAUAUCUUGGCAGCAGCUGGCUCUACAGACAUGUGCCAAGA-------GCCAACUGGCAA ................((((..((((........((((((((.((.(((......)))...))))))))))-------))))...)))). ( -26.30) >DroEre_CAF1 50733 83 + 1 AUUGUUAUUUUCUAGCUGCCUGUACUUGUAUACAUCUUGGCAGCAGCUGGGUCUACGGACAUGUGCCAAGA-------GCCAACCGCCAA ...........((((((((.(((...((....)).....)))))))))))(((....))).((.((.....-------))))........ ( -22.00) >DroYak_CAF1 54150 83 + 1 AUUGUUCUUUUUUAGCUGCCUGUAGUUGUAUAUAUCUUGGCAGCAGCUGGGUCUACGGACAUGUGCCAAGA-------GCCAACCGCCAA ...((((((.....((((((.(((........)))...)))))).((...(((....)))....)).))))-------)).......... ( -23.10) >consensus AUUGUUCUUUUUUAGCUGCCCGUAGUUGUAUAUAUCUUGGCAGCAGCUGGCUCUACAGACAUGUGCCAAGA_______GCCAACCGGCAA ................((((.((...........((((((((.((.(((......)))...))))))))))...........)).)))). (-19.35 = -19.91 + 0.56)


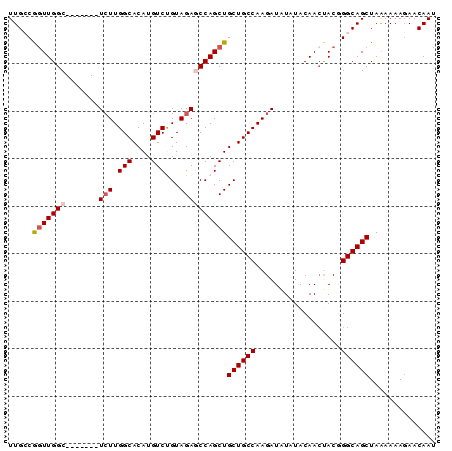
| Location | 11,713,520 – 11,713,610 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -17.62 |
| Energy contribution | -18.26 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11713520 90 - 27905053 UUGCCGGUUGGCUCUUGGCUGUUGGCACAUGUCUGUAGAGCCAGCUGCUGCCAAGAUAUAUACAACUACGGGCAGCUAAAAAAGAACAAU .....((((((((((.((((((....))).)))...))))))))))((((((.((..........))...)))))).............. ( -30.10) >DroSec_CAF1 50809 90 - 1 UUGCCGCUUGGCUCUUGGCUCUUGGCACAUGUCUGUAGAGCCAGCUGCUGCCAAGAUAUAUACAACCACGGGCAGCUAAAAAAGAACAAU ..(((....)))((((((((((.(((....)))...))))))....((((((..................)))))).....))))..... ( -28.07) >DroSim_CAF1 52483 83 - 1 UUGCCAGUUGGC-------UCUUGGCACAUGUCUGUAGAGCCAGCUGCUGCCAAGAUAUAUACAACCACGGGCAGCUAAAAAAGAACAAU .....(((((((-------(((.(((....)))...))))))))))((((((..................)))))).............. ( -29.07) >DroEre_CAF1 50733 83 - 1 UUGGCGGUUGGC-------UCUUGGCACAUGUCCGUAGACCCAGCUGCUGCCAAGAUGUAUACAAGUACAGGCAGCUAGAAAAUAACAAU (((..((((((.-------(((((((....).))).))).))))))((((((....(((((....)))))))))))..........))). ( -26.10) >DroYak_CAF1 54150 83 - 1 UUGGCGGUUGGC-------UCUUGGCACAUGUCCGUAGACCCAGCUGCUGCCAAGAUAUAUACAACUACAGGCAGCUAAAAAAGAACAAU (((..((((((.-------(((((((....).))).))).))))))((((((..................))))))..........))). ( -22.67) >consensus UUGCCGGUUGGC_______UCUUGGCACAUGUCUGUAGAGCCAGCUGCUGCCAAGAUAUAUACAACUACGGGCAGCUAAAAAAGAACAAU .....(((((((.......(((.(((....)))...))))))))))((((((..................)))))).............. (-17.62 = -18.26 + 0.64)


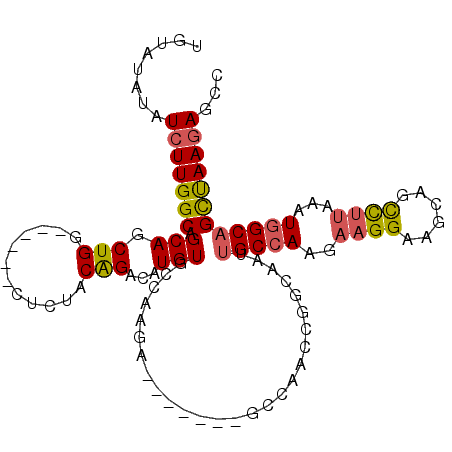
| Location | 11,713,546 – 11,713,650 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.88 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.93 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11713546 104 + 27905053 UGUAUAUAUCUUGGCAGCAGCUGG------CUCUACAGACAUGUGCCAACAGCCAAGAGCCAACCGGCAAUUGCCAAGAAGGAAGCAGCCUUAAAUUGCAGCUAAAAGCC ........(((((((((..(((((------((((...(...(((....))).)..))))))....)))..)))))))))....(((.((........)).)))....... ( -29.80) >DroSec_CAF1 50835 104 + 1 UGUAUAUAUCUUGGCAGCAGCUGG------CUCUACAGACAUGUGCCAAGAGCCAAGAGCCAAGCGGCAAUUGCCAAGAAGGAAGCAGCCUUAAAUGGCAGCUAAGAGCC ........(((((((.((.(((((------(((((((....)))((.....))..)))))).))).))...(((((..((((......))))...))))))))))))... ( -38.50) >DroSim_CAF1 52509 97 + 1 UGUAUAUAUCUUGGCAGCAGCUGG------CUCUACAGACAUGUGCCAAGA-------GCCAACUGGCAAUUGCCAAGAAGGAAGCAGCCUUAAAUGGCAGCUAAGAGCC ........(((((((..(((.(((------(((((((....)))....)))-------)))).))).....(((((..((((......))))...))))))))))))... ( -36.00) >DroEre_CAF1 50759 97 + 1 UGUAUACAUCUUGGCAGCAGCUGG------GUCUACGGACAUGUGCCAAGA-------GCCAACCGCCAAAAGCCAAGAUGGAAGCAGCCUUAAAUGGCAGUUAAGAGCC (((...(((((((((....((.((------(((....))).((.((.....-------)))).)))).....)))))))))...)))(((......)))........... ( -31.50) >DroYak_CAF1 54176 97 + 1 UGUAUAUAUCUUGGCAGCAGCUGG------GUCUACGGACAUGUGCCAAGA-------GCCAACCGCCAAUUGCCAACAAGGAAGCAGCCUUAAAUGGCAGCUAAGAGUC ........(((((((.((.(.(((------.(((..((.(....))).)))-------.))).).))....(((((..((((......))))...))))))))))))... ( -29.60) >DroAna_CAF1 50408 89 + 1 UGCAUAUAUCUUGGCAGCAGCUGCUGGAAUGUCCACGGACAUGUGGC---------------------AAGUGCCAAGAAGGAAGCGCUUUGAAUUGGCAGCCAAGAGCC ........(((((((....((..(....(((((....))))))..))---------------------...((((((.((((.....))))...)))))))))))))... ( -31.40) >consensus UGUAUAUAUCUUGGCAGCAGCUGG______CUCUACAGACAUGUGCCAAGA_______GCCAACCGGCAAUUGCCAAGAAGGAAGCAGCCUUAAAUGGCAGCUAAGAGCC ........(((((((.(((.(((............)))...)))...........................(((((..((((......))))...))))))))))))... (-19.90 = -19.93 + 0.03)

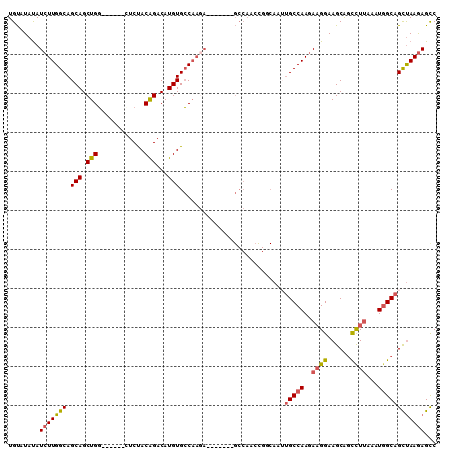
| Location | 11,713,546 – 11,713,650 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.88 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -17.81 |
| Energy contribution | -20.23 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11713546 104 - 27905053 GGCUUUUAGCUGCAAUUUAAGGCUGCUUCCUUCUUGGCAAUUGCCGGUUGGCUCUUGGCUGUUGGCACAUGUCUGUAGAG------CCAGCUGCUGCCAAGAUAUAUACA ((.....(((.((........)).))).)).((((((((...((.((((((((((.((((((....))).)))...))))------))))))))))))))))........ ( -38.40) >DroSec_CAF1 50835 104 - 1 GGCUCUUAGCUGCCAUUUAAGGCUGCUUCCUUCUUGGCAAUUGCCGCUUGGCUCUUGGCUCUUGGCACAUGUCUGUAGAG------CCAGCUGCUGCCAAGAUAUAUACA ((.....(((.(((......))).))).)).((((((((...((.(((.((((((.(((...........)))...))))------))))).))))))))))........ ( -38.80) >DroSim_CAF1 52509 97 - 1 GGCUCUUAGCUGCCAUUUAAGGCUGCUUCCUUCUUGGCAAUUGCCAGUUGGC-------UCUUGGCACAUGUCUGUAGAG------CCAGCUGCUGCCAAGAUAUAUACA ((.....(((.(((......))).))).)).((((((((...((.(((((((-------(((.(((....)))...))))------))))))))))))))))........ ( -41.80) >DroEre_CAF1 50759 97 - 1 GGCUCUUAACUGCCAUUUAAGGCUGCUUCCAUCUUGGCUUUUGGCGGUUGGC-------UCUUGGCACAUGUCCGUAGAC------CCAGCUGCUGCCAAGAUGUAUACA (((........)))...............(((((((((....(((((((((.-------(((((((....).))).))).------))))))))))))))))))...... ( -37.90) >DroYak_CAF1 54176 97 - 1 GACUCUUAGCUGCCAUUUAAGGCUGCUUCCUUGUUGGCAAUUGGCGGUUGGC-------UCUUGGCACAUGUCCGUAGAC------CCAGCUGCUGCCAAGAUAUAUACA ...((((.(((((((..(((((......))))).)))))...(((((((((.-------(((((((....).))).))).------))))))))))).))))........ ( -34.80) >DroAna_CAF1 50408 89 - 1 GGCUCUUGGCUGCCAAUUCAAAGCGCUUCCUUCUUGGCACUU---------------------GCCACAUGUCCGUGGACAUUCCAGCAGCUGCUGCCAAGAUAUAUGCA (((........)))..........((.....((((((((.((---------------------((...(((((....)))))....))))....)))))))).....)). ( -27.80) >consensus GGCUCUUAGCUGCCAUUUAAGGCUGCUUCCUUCUUGGCAAUUGCCGGUUGGC_______UCUUGGCACAUGUCCGUAGAC______CCAGCUGCUGCCAAGAUAUAUACA .......(((.(((......))).)))....((((((((.....(((((((............(((....))).............))))))).))))))))........ (-17.81 = -20.23 + 2.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:18 2006