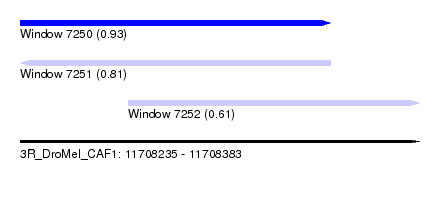
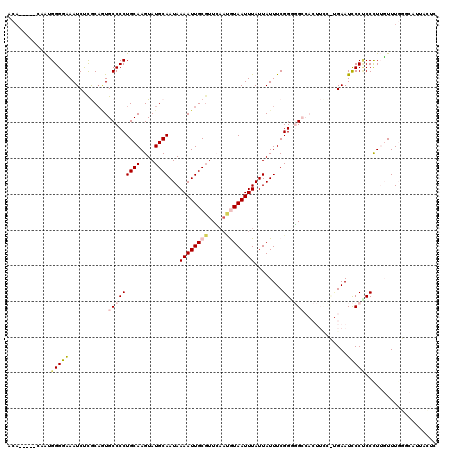
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,708,235 – 11,708,383 |
| Length | 148 |
| Max. P | 0.925725 |

| Location | 11,708,235 – 11,708,350 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -18.97 |
| Energy contribution | -18.25 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
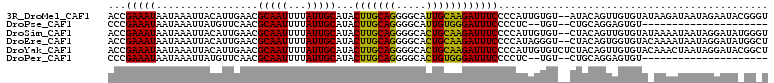
>3R_DroMel_CAF1 11708235 115 + 27905053 GAGUAAUGCCCAAACAAGGGAGGGAUUCAGGGAAGUGGCCACCGAAAUAAUAAAUUACAUUGAACGCAAUUUUAUUGCAUACUUGCAGGGGCAUUGCAAGAUUUCCCCAUUG-----UGU ((((....(((......)))....)))).(((((((.............................(((((...)))))...(((((((.....)))))))))))))).....-----... ( -28.30) >DroPse_CAF1 50289 96 + 1 GUGG-------AAAAUAGGGGGGAAUUC----------CGCCCGAAAUAAUAAAUUAUGUUCAACGCAAUUUUAUUGCAUACUUGCAGGGGCAUUGUGGGAUUUCCCCUC-------UGU ....-------...(((((((((((.((----------(....(((((((....)))).)))..((((((.(..(((((....)))))..).))))))))).))))))))-------))) ( -34.40) >DroSim_CAF1 47191 115 + 1 GAGUAAUGCCCAAACAAGGGAGGGAUUCAAGGAAGUGGCCACCGAAAUAAUAAAUUACAUUGAACGCAAUUUUAUUGCAUACUUGCAGGGGCACUGCAAGAUUUCCCCAUUG-----UGU ((((....(((......)))....))))..((((((.............................(((((...)))))...(((((((.....)))))))))))))......-----... ( -28.00) >DroEre_CAF1 45508 115 + 1 GAGUAAUGCCCAAACAAGGGUGGGAUUCAAGGAAGUGGCCACCGAAAUAAUAAAUUACAUUGAACGCAAUUUUAUUGCAUACUUGCAGGGGCACUGCAAGAUUUCCCCAUAG-----GGU ..(((.(((((...(((((((((..(((...)))....)))))......................(((((...)))))...))))...))))).))).......(((....)-----)). ( -30.80) >DroAna_CAF1 45372 120 + 1 GACUAAUGGGCAAACAAAGGAGGGAUUCGGAAAAGUGGCCCCCGAAAUAAUAAAUUACUGGGAGGGCAAUUUUAUUGCAUACUUGCAGGGGCAAGGCGAGAUUCCCCCGUUUGCAUGUGG ..(((....((((((...((.((((.((...(((((.((((((.(.............).)).)))).))))).((((...(((((....))))))))))).))))))))))))...))) ( -37.52) >DroPer_CAF1 46181 96 + 1 GUGG-------AAAAUAGGGGGGAAUUC----------CACCCGAAAUAAUAAAUUAUGUUCAACGCAAUUUUAUUGCAUACUUGCAGGGGCACUGUGGGAUUUCCCCUC-------UGU ....-------...(((((((((((.((----------(....(((((((....)))).)))..((((......(((((....)))))......))))))).))))))))-------))) ( -31.60) >consensus GAGUAAUGCCCAAACAAGGGAGGGAUUCA_GGAAGUGGCCACCGAAAUAAUAAAUUACAUUGAACGCAAUUUUAUUGCAUACUUGCAGGGGCACUGCAAGAUUUCCCCAUUG_____UGU .................(((.(((((.......................................(((((...)))))...(((((((.....)))))))))))))))............ (-18.97 = -18.25 + -0.72)

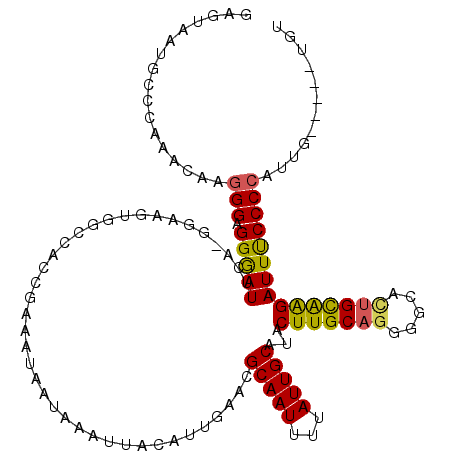
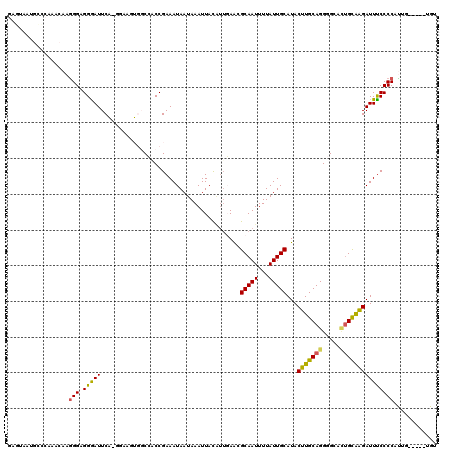
| Location | 11,708,235 – 11,708,350 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -13.28 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
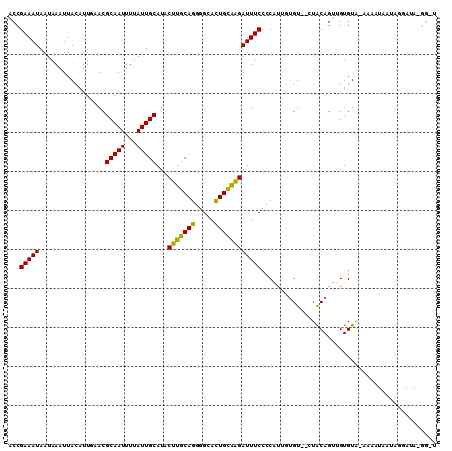
>3R_DroMel_CAF1 11708235 115 - 27905053 ACA-----CAAUGGGGAAAUCUUGCAAUGCCCCUGCAAGUAUGCAAUAAAAUUGCGUUCAAUGUAAUUUAUUAUUUCGGUGGCCACUUCCCUGAAUCCCUCCCUUGUUUGGGCAUUACUC ...-----.................(((((((..(((((.........(((((((((...))))))))).....(((((.((......))))))).......)))))..))))))).... ( -26.70) >DroPse_CAF1 50289 96 - 1 ACA-------GAGGGGAAAUCCCACAAUGCCCCUGCAAGUAUGCAAUAAAAUUGCGUUGAACAUAAUUUAUUAUUUCGGGCG----------GAAUUCCCCCCUAUUUU-------CCAC ..(-------(.((((((.(((......((((........(((((((...))))))).(((.((((....))))))))))))----------)).)))))).)).....-------.... ( -25.60) >DroSim_CAF1 47191 115 - 1 ACA-----CAAUGGGGAAAUCUUGCAGUGCCCCUGCAAGUAUGCAAUAAAAUUGCGUUCAAUGUAAUUUAUUAUUUCGGUGGCCACUUCCUUGAAUCCCUCCCUUGUUUGGGCAUUACUC ...-----.((((((((..((..(.((((((((((((....))))...(((((((((...)))))))))........)).)).))))..)..)).)))).(((......))))))).... ( -27.70) >DroEre_CAF1 45508 115 - 1 ACC-----CUAUGGGGAAAUCUUGCAGUGCCCCUGCAAGUAUGCAAUAAAAUUGCGUUCAAUGUAAUUUAUUAUUUCGGUGGCCACUUCCUUGAAUCCCACCCUUGUUUGGGCAUUACUC .((-----(....))).........(((((((..(((((.........(((((((((...)))))))))........(((((...(......)....))))))))))..))))))).... ( -32.00) >DroAna_CAF1 45372 120 - 1 CCACAUGCAAACGGGGGAAUCUCGCCUUGCCCCUGCAAGUAUGCAAUAAAAUUGCCCUCCCAGUAAUUUAUUAUUUCGGGGGCCACUUUUCCGAAUCCCUCCUUUGUUUGCCCAUUAGUC ......((((((((((((.((..(((((((....)))))...)).........((((((..(((((....)))))..)))))).........)).))))))....))))))......... ( -33.90) >DroPer_CAF1 46181 96 - 1 ACA-------GAGGGGAAAUCCCACAGUGCCCCUGCAAGUAUGCAAUAAAAUUGCGUUGAACAUAAUUUAUUAUUUCGGGUG----------GAAUUCCCCCCUAUUUU-------CCAC ..(-------(.((((((.(((......((((........(((((((...))))))).(((.((((....))))))))))))----------)).)))))).)).....-------.... ( -24.20) >consensus ACA_____CAAUGGGGAAAUCUCGCAGUGCCCCUGCAAGUAUGCAAUAAAAUUGCGUUCAAUGUAAUUUAUUAUUUCGGGGGCCACUUCC_UGAAUCCCUCCCUUGUUUGGGCAUUACUC ............(((((...........(((((((((....))))...((((((((.....))))))))........)).)))............))))).................... (-13.28 = -13.92 + 0.64)

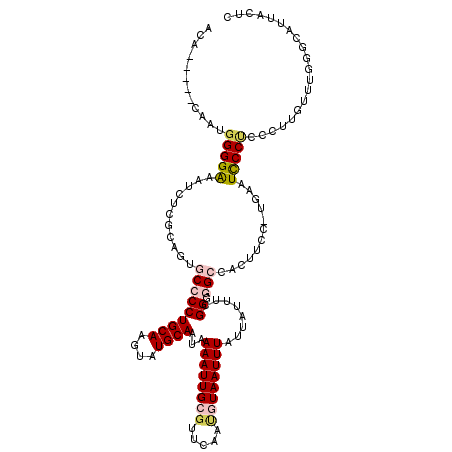
| Location | 11,708,275 – 11,708,383 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.15 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -15.45 |
| Energy contribution | -14.57 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
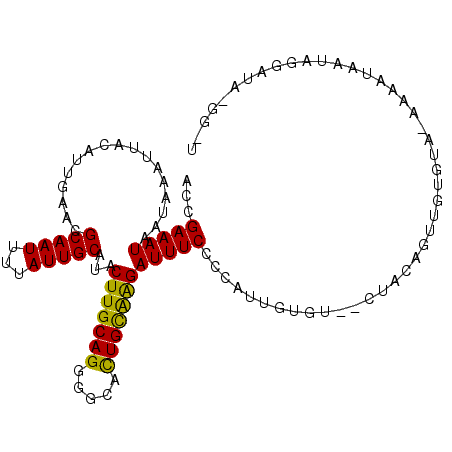
>3R_DroMel_CAF1 11708275 108 + 27905053 ACCGAAAUAAUAAAUUACAUUGAACGCAAUUUUAUUGCAUACUUGCAGGGGCAUUGCAAGAUUUCCCCAUUGUGU--AUACAGUUGUGUAUAAGAUAAUAGAAUACGGGU .(((...........((((.((...(((((...)))))...(((((((.....))))))).......)).))))(--(((((....)))))).............))).. ( -22.30) >DroPse_CAF1 50312 85 + 1 CCCGAAAUAAUAAAUUAUGUUCAACGCAAUUUUAUUGCAUACUUGCAGGGGCAUUGUGGGAUUUCCCCUC--UGU--CUGCAGGAGUGU--------------------- ........((((((...(((.....)))..))))))((((.(((((((..(((..(.(((....))))..--)))--))))))).))))--------------------- ( -21.60) >DroSim_CAF1 47231 108 + 1 ACCGAAAUAAUAAAUUACAUUGAACGCAAUUUUAUUGCAUACUUGCAGGGGCACUGCAAGAUUUCCCCAUUGUGU--CUACAGUUGUGUAUAAAAUAAUAGGAUAUGGGU ...(((((.................(((((...)))))...(((((((.....))))))))))))(((((....(--(((..(((........)))..))))..))))). ( -25.40) >DroEre_CAF1 45548 108 + 1 ACCGAAAUAAUAAAUUACAUUGAACGCAAUUUUAUUGCAUACUUGCAGGGGCACUGCAAGAUUUCCCCAUAGGGU--CUACAGUGGUGUACAAAAUAAUAGGAUAUGGCU ...(((((.................(((((...)))))...(((((((.....)))))))))))).(((((...(--(((...((.....))......)))).))))).. ( -23.00) >DroYak_CAF1 45880 110 + 1 ACCGAAAUAAUAAAUUACAUUGAACGCAAUUUUAUUGCAUACUUGCAGGGGCACUGCAAGAUUUCCCCAUUGUGUCUCUACAGUUGUGUACAAACUAAUAGGAUACGGCU .(((((((.................(((((...)))))...(((((((.....))))))))))))......(((((.(((.((((.......))))..)))))))))).. ( -26.90) >DroPer_CAF1 46204 85 + 1 CCCGAAAUAAUAAAUUAUGUUCAACGCAAUUUUAUUGCAUACUUGCAGGGGCACUGUGGGAUUUCCCCUC--UGU--CUGCAGGAGUGU--------------------- ........((((((...(((.....)))..))))))((((.(((((((..(((..(.(((....))))..--)))--))))))).))))--------------------- ( -21.60) >consensus ACCGAAAUAAUAAAUUACAUUGAACGCAAUUUUAUUGCAUACUUGCAGGGGCACUGCAAGAUUUCCCCAUUGUGU__CUACAGUUGUGUA_AAAAUAAUAGGAUA_GG_U ...(((((.................(((((...)))))...(((((((.....))))))))))))............................................. (-15.45 = -14.57 + -0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:13 2006