
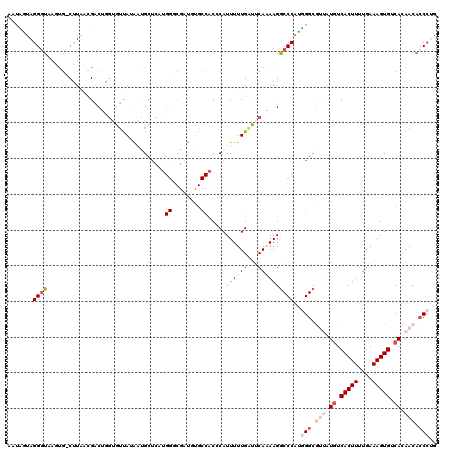
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,703,927 – 11,704,115 |
| Length | 188 |
| Max. P | 0.999919 |

| Location | 11,703,927 – 11,704,047 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.98 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -11.34 |
| Energy contribution | -13.88 |
| Covariance contribution | 2.54 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.32 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11703927 120 + 27905053 AAUCAUAGGGUACGUGUCUUAACGCCUGUUGUUAUAAUGCUCAUGGCCGAUGUGCCACCCAUUUUUGAUUCAAAAGACCCAUGGGCGUUAUUUCACUUUUGAAAGUGUCACAACACCCUG .....((((((..((((....))))..(((((.(((((((((((((.(((.(((.....)))..)))..((....)).)))))))))))))..(((((....)))))..))))))))))) ( -32.60) >DroSec_CAF1 41241 119 + 1 AAUUGUAGGGUAAGUG-CUUGGUGACUGGAGUUAAAUUGCUCCUGGACGAUGCGCCACCCAUUUUUGAUUCAAAAGGCCCAUGGGCGUUAUGUCACUUUUGAAAGUGUCACAACACCCUG .......((((..(((-(.(.((..(.(((((......))))).).)).).))))......((((((...))))))))))..(((.(((.((.(((((....))))).)).))).))).. ( -35.80) >DroSim_CAF1 42837 119 + 1 AAUUGUAGGGUAAGUG-CCUGGUGACUGGAGUUAAAUUGCUCCUGGGCGAUGUGCCACCCAUUUUUGAUUCAAAAGGCCCAUGGGCGUUAUGUCACUUUUGAAAGUGUCACAACACCCUG .....((((((....(-((((....).(((((......))))).))))(.((((.(((.((....)).(((((((((((....)))(......).)))))))).))).)))).))))))) ( -40.10) >DroEre_CAF1 40980 115 + 1 AAUAGCAGGGUAAACGUCUUAACGACAAGUUUUAUAACGCGCACGGGCGUUAUGCCACACAUUUUUGUGUCAUAAGGCCCACUGGCG--CUGUCACUUUUGAAAGUGUCAUAAGUGC--- ....((.........(((.....)))..(((....)))))((((((((.(((((.((((......)))).))))).))))..(((((--((.(((....))).)))))))...))))--- ( -39.50) >DroYak_CAF1 41023 103 + 1 AACAGUAGGUU-ACCAACUUAACGUCAAGUUGCAUGACGCU----------------CACAUUUUUGUGGCAUAAGGCCCACUGGCAAAAUGUCACUUUUGAAAGUGUCAUAACACCCAG .......(((.-..((((((......)))))).((((((((----------------..((.....(((((((...(((....)))...)))))))...))..))))))))...)))... ( -29.50) >consensus AAUAGUAGGGUAAGUG_CUUAACGACUGGUGUUAUAAUGCUCAUGGGCGAUGUGCCACCCAUUUUUGAUUCAAAAGGCCCAUGGGCGUUAUGUCACUUUUGAAAGUGUCACAACACCCUG .......((((.................................((........)).....((((((...))))))))))..(((.(((.((.(((((....))))).)).))).))).. (-11.34 = -13.88 + 2.54)


| Location | 11,703,927 – 11,704,047 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.98 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -16.30 |
| Energy contribution | -19.84 |
| Covariance contribution | 3.54 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.45 |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
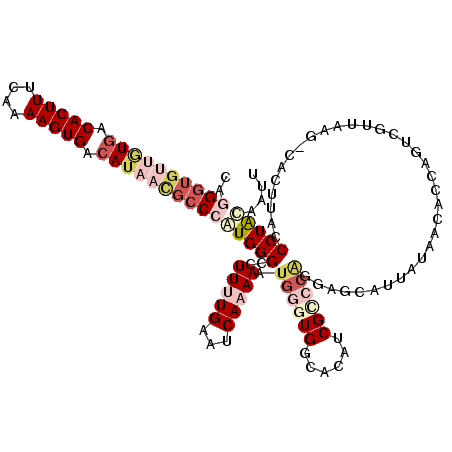
>3R_DroMel_CAF1 11703927 120 - 27905053 CAGGGUGUUGUGACACUUUCAAAAGUGAAAUAACGCCCAUGGGUCUUUUGAAUCAAAAAUGGGUGGCACAUCGGCCAUGAGCAUUAUAACAACAGGCGUUAAGACACGUACCCUAUGAUU ..(((((((((..(((((....)))))..)))))))))..((((((.(((..(((....)))(((((......)))))...........))).))((((......))))))))....... ( -33.90) >DroSec_CAF1 41241 119 - 1 CAGGGUGUUGUGACACUUUCAAAAGUGACAUAACGCCCAUGGGCCUUUUGAAUCAAAAAUGGGUGGCGCAUCGUCCAGGAGCAAUUUAACUCCAGUCACCAAG-CACUUACCCUACAAUU ..((((((((((.(((((....))))).))))))))))..(((..(((((...)))))...(((((((.......).((((........)))).))))))...-......)))....... ( -37.40) >DroSim_CAF1 42837 119 - 1 CAGGGUGUUGUGACACUUUCAAAAGUGACAUAACGCCCAUGGGCCUUUUGAAUCAAAAAUGGGUGGCACAUCGCCCAGGAGCAAUUUAACUCCAGUCACCAGG-CACUUACCCUACAAUU ..((((((((((.(((((....))))).))))))))))....((((..(((........(((((((....)))))))((((........))))..)))..)))-)............... ( -44.00) >DroEre_CAF1 40980 115 - 1 ---GCACUUAUGACACUUUCAAAAGUGACAG--CGCCAGUGGGCCUUAUGACACAAAAAUGUGUGGCAUAACGCCCGUGCGCGUUAUAAAACUUGUCGUUAAGACGUUUACCCUGCUAUU ---(((.(((((((((((....))))....(--(((...(((((.(((((.((((......)))).))))).))))).))))))))))).....(((.....)))........))).... ( -35.40) >DroYak_CAF1 41023 103 - 1 CUGGGUGUUAUGACACUUUCAAAAGUGACAUUUUGCCAGUGGGCCUUAUGCCACAAAAAUGUG----------------AGCGUCAUGCAACUUGACGUUAAGUUGGU-AACCUACUGUU ...((((..(((.(((((....))))).)))..))))((((((....((((((((....))))----------------.))))....(((((((....)))))))..-..))))))... ( -30.30) >consensus CAGGGUGUUGUGACACUUUCAAAAGUGACAUAACGCCCAUGGGCCUUUUGAAUCAAAAAUGGGUGGCACAUCGCCCAGGAGCAUUAUAACACCAGUCGUUAAG_CACUUACCCUACAAUU ..((((((((((.(((((....))))).))))))))))(((((..(((((...))))).((((((......))))))..................................))))).... (-16.30 = -19.84 + 3.54)

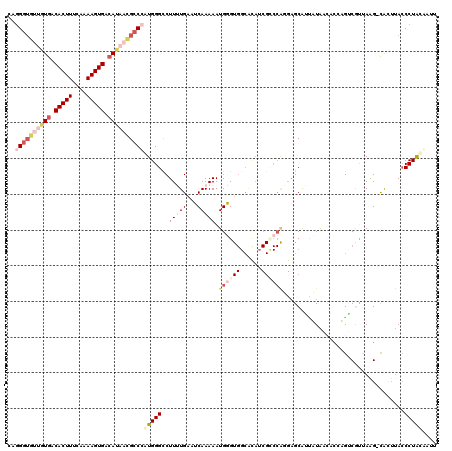
| Location | 11,703,967 – 11,704,080 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -22.80 |
| Energy contribution | -24.17 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11703967 113 - 27905053 ACUUAACUUGAAGAGCAAUCGAUAAUUAGUCAGCAGGGUGUUGUGACACUUUCAAAAGUGAAAUAACGCCCAUGGGUCUUUUGAAUCAAAAAUGGGUGGCACAUCGGCCAUGA .........(((((.(....(((.....)))....(((((((((..(((((....)))))..)))))))))...).)))))..............(((((......))))).. ( -30.90) >DroSec_CAF1 41280 113 - 1 ACUUAAUUCAAAGAGCAAUCGAUAAUUAGCCUGCAGGGUGUUGUGACACUUUCAAAAGUGACAUAACGCCCAUGGGCCUUUUGAAUCAAAAAUGGGUGGCGCAUCGUCCAGGA .....(((((((((....))........((((...((((((((((.(((((....))))).))))))))))..))))..)))))))......(((..((....))..)))... ( -35.90) >DroSim_CAF1 42876 113 - 1 AAUUAAUUUAAAGAGCAAUCGAUAAUUAGCCUGCAGGGUGUUGUGACACUUUCAAAAGUGACAUAACGCCCAUGGGCCUUUUGAAUCAAAAAUGGGUGGCACAUCGCCCAGGA .....(((((((((....))........((((...((((((((((.(((((....))))).))))))))))..))))..)))))))......(((((((....)))))))... ( -38.50) >DroYak_CAF1 41062 97 - 1 ACUUACCUCAAGGCGCAGGCCAAAAUUAGCCUUCUGGGUGUUAUGACACUUUCAAAAGUGACAUUUUGCCAGUGGGCCUUAUGCCACAAAAAUGUG----------------A .........(((((.(((((........))).....((((..(((.(((((....))))).)))..))))..)).)))))....((((....))))----------------. ( -26.70) >consensus ACUUAACUCAAAGAGCAAUCGAUAAUUAGCCUGCAGGGUGUUGUGACACUUUCAAAAGUGACAUAACGCCCAUGGGCCUUUUGAAUCAAAAAUGGGUGGCACAUCG_CCAGGA .......(((((((....))........((((...((((((((((.(((((....))))).))))))))))..))))..)))))........(((((((....)))))))... (-22.80 = -24.17 + 1.38)



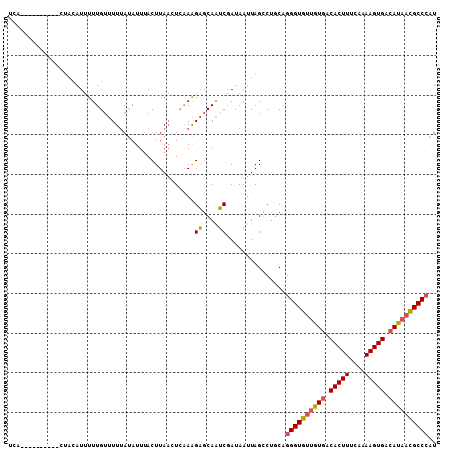
| Location | 11,704,007 – 11,704,115 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.38 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -16.95 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.72 |
| SVM decision value | 4.55 |
| SVM RNA-class probability | 0.999919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11704007 108 - 27905053 UCAUAAAUUACUGCUACAUUUUUGAUUUUAUAUUUACUUAACUUGAAGAGCAAUCGAUAAUUAGUCAGCAGGGUGUUGUGACACUUUCAAAAGUGAAAUAACGCCCAU ..........((((((.(((.((((((.....((((.......))))....)))))).)))))).)))..(((((((((..(((((....)))))..))))))))).. ( -25.90) >DroSec_CAF1 41320 92 - 1 ----------------AAUUUUUGUUUUUAUAUUUACUUAAUUCAAAGAGCAAUCGAUAAUUAGCCUGCAGGGUGUUGUGACACUUUCAAAAGUGACAUAACGCCCAU ----------------..((((((...(((........)))..))))))(((..(........)..))).((((((((((.(((((....))))).)))))))))).. ( -25.90) >DroYak_CAF1 41086 99 - 1 UCA---------CCUACAUGUUUGUUUUAACAUUAACUUACCUCAAGGCGCAGGCCAAAAUUAGCCUUCUGGGUGUUAUGACACUUUCAAAAGUGACAUUUUGCCAGU ...---------(((....((..(((........)))..))....))).(.((((........)))).)..((((..(((.(((((....))))).)))..))))... ( -18.90) >consensus UCA__________CUACAUUUUUGUUUUUAUAUUUACUUAACUCAAAGAGCAAUCGAUAAUUAGCCUGCAGGGUGUUGUGACACUUUCAAAAGUGACAUAACGCCCAU ...............................................((....))...............((((((((((.(((((....))))).)))))))))).. (-16.95 = -17.40 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:08 2006