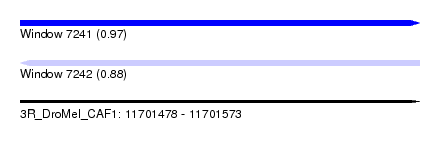
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,701,478 – 11,701,573 |
| Length | 95 |
| Max. P | 0.969843 |

| Location | 11,701,478 – 11,701,573 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 68.39 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -15.84 |
| Energy contribution | -16.57 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11701478 95 + 27905053 ACGCCCGCAUCCGAAUCCGAAUCCAAAUCGGAUU-CGCAGGCAGGCGG----UCGUCGGCGAUUCC-GCCGCUCAAGUUGAACGCAGCAGUCGGCGGCGGC .((((.((...(((((((((.......)))))))-))...)).))))(----(((....)))).((-((((((...((((....))))....)))))))). ( -45.80) >DroSec_CAF1 37630 77 + 1 ACGCCCGCAUCCGAAUCCGAAUCCAAAUCGGAUU-CGAA----GGCGG----UCGUCGCCGAUUCC-GCCGCUCAA----------GCAGUCGGCGG---- (((.((((...(((((((((.......)))))))-))..----.))))----.)))((((((((..-((.......----------)))))))))).---- ( -36.20) >DroSim_CAF1 40456 81 + 1 ACGCCCGCAUCCGAAUCCGAAUCCAAAUCGGAUU-CGAA----GGCGG----UCGUCGUCGAUUCC-GCCGCUCAA----------GCAGUCGGCGGCGGC (((.((((...(((((((((.......)))))))-))..----.))))----.)))........((-((((((...----------......)))))))). ( -38.20) >DroEre_CAF1 38575 81 + 1 ACGCCCGCAUCCGA------AUCCAAAUCGGAUU-CGCA----GGCGC----UCGUCGGCGAUUCC-GCCGCUGAAGUCGGACGCAGCCGUCGGCGG---- .((((.((...(((------((((.....)))))-))..----(((..----.((((((((....)-)))((....))..))))..))))).)))).---- ( -35.80) >DroYak_CAF1 38470 90 + 1 ACGUCCGCAUCCGAAUCCGAAUCCAAGUCGGAUU-CGCA----GGCGGUCGGUCGUCGGCGAUUCC-GCCGCUGAAGUCGGACGCAGCCGUCGGCG----- .((.((((...(((((((((.......)))))))-))..----.)))).))((((.((((...(((-(.(......).))))....)))).)))).----- ( -44.90) >DroAna_CAF1 39023 76 + 1 ACGGGCGAUUCGGAGGCAGCGCCCGACGACGAUGCCGCU----GCCGA----CUGUGGGCGGCUACGGCUGCUA-----------------CGACGGCGAC .((((((...(....)...)))))).............(----((((.----.(((((.((((....)))))))-----------------)).))))).. ( -31.30) >consensus ACGCCCGCAUCCGAAUCCGAAUCCAAAUCGGAUU_CGCA____GGCGG____UCGUCGGCGAUUCC_GCCGCUCAA__________GCAGUCGGCGG____ .((((.((((((((.............))))))..........(((((....(((....)))......)))))................)).))))..... (-15.84 = -16.57 + 0.73)

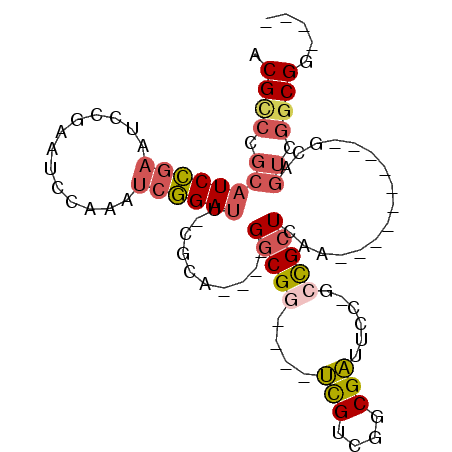

| Location | 11,701,478 – 11,701,573 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 68.39 |
| Mean single sequence MFE | -40.43 |
| Consensus MFE | -12.03 |
| Energy contribution | -13.04 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11701478 95 - 27905053 GCCGCCGCCGACUGCUGCGUUCAACUUGAGCGGC-GGAAUCGCCGACGA----CCGCCUGCCUGCG-AAUCCGAUUUGGAUUCGGAUUCGGAUGCGGGCGU ..((.((.((((((((((...........)))))-))..))).)).)).----.(((((((.(.((-(((((((.......))))))))).).))))))). ( -45.60) >DroSec_CAF1 37630 77 - 1 ----CCGCCGACUGC----------UUGAGCGGC-GGAAUCGGCGACGA----CCGCC----UUCG-AAUCCGAUUUGGAUUCGGAUUCGGAUGCGGGCGU ----.((((((((((----------(.....)))-))..))))))(((.----((((.----((((-(((((((.......))))))))))).)))).))) ( -42.90) >DroSim_CAF1 40456 81 - 1 GCCGCCGCCGACUGC----------UUGAGCGGC-GGAAUCGACGACGA----CCGCC----UUCG-AAUCCGAUUUGGAUUCGGAUUCGGAUGCGGGCGU .((((((((((....----------))).)))))-))........(((.----((((.----((((-(((((((.......))))))))))).)))).))) ( -43.70) >DroEre_CAF1 38575 81 - 1 ----CCGCCGACGGCUGCGUCCGACUUCAGCGGC-GGAAUCGCCGACGA----GCGCC----UGCG-AAUCCGAUUUGGAU------UCGGAUGCGGGCGU ----......(((.((((((((((.(((((...(-(((.((((.(.(..----..).)----.)))-).))))..))))).------)))))))))).))) ( -38.90) >DroYak_CAF1 38470 90 - 1 -----CGCCGACGGCUGCGUCCGACUUCAGCGGC-GGAAUCGCCGACGACCGACCGCC----UGCG-AAUCCGACUUGGAUUCGGAUUCGGAUGCGGACGU -----.....(((.((((((((((.(((.(((((-((..(((........))))))))----.))(-(((((.....))))))))).)))))))))).))) ( -43.30) >DroAna_CAF1 39023 76 - 1 GUCGCCGUCG-----------------UAGCAGCCGUAGCCGCCCACAG----UCGGC----AGCGGCAUCGUCGUCGGGCGCUGCCUCCGAAUCGCCCGU .....((.((-----------------.....(((((.(((((.....)----.))))----.)))))..)).)).((((((.(........).)))))). ( -28.20) >consensus ____CCGCCGACUGC__________UUGAGCGGC_GGAAUCGCCGACGA____CCGCC____UGCG_AAUCCGAUUUGGAUUCGGAUUCGGAUGCGGGCGU .....((((....................((((......(((....)))....))))...........((((((.............))))))...)))). (-12.03 = -13.04 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:04 2006