
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,700,410 – 11,700,500 |
| Length | 90 |
| Max. P | 0.981357 |

| Location | 11,700,410 – 11,700,500 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 66.71 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -8.93 |
| Energy contribution | -10.90 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.25 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
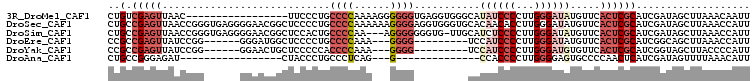
>3R_DroMel_CAF1 11700410 90 + 27905053 CUGUCGAGUUAAC-----------------UUCCCUGCCCCAAAAGGGGGGUGAGGUGGGCAUAUCCCCUUGGGAUAUGUUCACUCGCAUCGAUAGCUUAAACAAUU (((((((((..((-----------------((((((........))))))))(((.((((((((((((...))))))))))))))))).)))))))........... ( -39.60) >DroSec_CAF1 36496 107 + 1 CUGCCGAGUUAACCGGGUGAGGGGAACGGCUCCCCUGCCCCAAAAAAGGGGAGGUGGGUGCACAACACCUUGGGAUAUGUUCACUCGCAUCGAUAGCUUAAACCAUU .....((((((..((((((((..(((((..((((((.((((......))))))..(((((.....))))).))))..))))).))))).))).))))))........ ( -38.70) >DroSim_CAF1 39319 103 + 1 CUGCCGAGUUAACCGGGUGAGGGGAACGGCUCCACUGCCCCAA---AGGGGGGGUG-UUGCAUCUCCCCUUGGGAUAUGUUCACUCGCAUCGAUAGCUUAAACCAUU .....((((((..((((((((..(((((((......)))((.(---((((((((((-...))))))))))).))....)))).))))).))).))))))........ ( -41.90) >DroEre_CAF1 37437 89 + 1 CCGCCGAGUUAUCCGG------GGGAUGGCUCCCCUGCCCCAAA---GGGG---------UCCAUCCCCUUGGGAUAUGUUCACUCGCAUCGGCAGCUUAAACCAUU ..(((((..(((((((------(((((((..(((((.......)---))))---------.)))))))))..)))))(((......))))))))............. ( -41.40) >DroYak_CAF1 37317 89 + 1 CCGCCGAGUUAUCCGG------GGAACUGCUCCCCCACCCCAAA---GGGG---------UCCAUCCCCUUGGGAUGUGUUCACUCGCAUCGGUAGCUUACCCCAUU .....((((((((.((------(((.....)))))....((((.---((((---------.....))))))))((((((......))))))))))))))........ ( -34.90) >DroAna_CAF1 37750 73 + 1 CUGCCGGGAGAU-----------------CUACCCUGCCCUCAG---G--------------CCACCCCUUGGGGAGUGCCCCAACUCAUCGAUAGUUUUAAACAUU (((.((((((..-----------------....((((....)))---)--------------.......((((((....))))))))).))).)))........... ( -18.10) >consensus CUGCCGAGUUAACCGG______GGAACGGCUCCCCUGCCCCAAA___GGGG_________CACAUCCCCUUGGGAUAUGUUCACUCGCAUCGAUAGCUUAAACCAUU ..(.(((((............................((((......))))...........((((((...)))))).....))))))................... ( -8.93 = -10.90 + 1.97)


| Location | 11,700,410 – 11,700,500 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 66.71 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -12.26 |
| Energy contribution | -12.93 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11700410 90 - 27905053 AAUUGUUUAAGCUAUCGAUGCGAGUGAACAUAUCCCAAGGGGAUAUGCCCACCUCACCCCCCUUUUGGGGCAGGGAA-----------------GUUAACUCGACAG ...((((...((.......))((((.((((((((((...))))))).....(((..((((......))))..)))..-----------------))).)))))))). ( -28.40) >DroSec_CAF1 36496 107 - 1 AAUGGUUUAAGCUAUCGAUGCGAGUGAACAUAUCCCAAGGUGUUGUGCACCCACCUCCCCUUUUUUGGGGCAGGGGAGCCGUUCCCCUCACCCGGUUAACUCGGCAG .(((((....)))))...(((((((((((...((((..((((.....))))...((((((......)))).))))))...)))).((......))...)))).))). ( -33.90) >DroSim_CAF1 39319 103 - 1 AAUGGUUUAAGCUAUCGAUGCGAGUGAACAUAUCCCAAGGGGAGAUGCAA-CACCCCCCCU---UUGGGGCAGUGGAGCCGUUCCCCUCACCCGGUUAACUCGGCAG .(((((....)))))...(((((((.(((........(((((((..((..-(((..((((.---..))))..)))..))..)))))))......))).)))).))). ( -35.34) >DroEre_CAF1 37437 89 - 1 AAUGGUUUAAGCUGCCGAUGCGAGUGAACAUAUCCCAAGGGGAUGGA---------CCCC---UUUGGGGCAGGGGAGCCAUCCC------CCGGAUAACUCGGCGG ...........(((((((.....(....).(((((...((((((((.---------((((---(.......)))))..)))))))------).)))))..))))))) ( -39.90) >DroYak_CAF1 37317 89 - 1 AAUGGGGUAAGCUACCGAUGCGAGUGAACACAUCCCAAGGGGAUGGA---------CCCC---UUUGGGGUGGGGGAGCAGUUCC------CCGGAUAACUCGGCGG ..............(((...(((((...(.((((((((((((.....---------.)))---)))))))))((((((...))))------)))....))))).))) ( -40.10) >DroAna_CAF1 37750 73 - 1 AAUGUUUAAAACUAUCGAUGAGUUGGGGCACUCCCCAAGGGGUGG--------------C---CUGAGGGCAGGGUAG-----------------AUCUCCCGGCAG ..((((................(..((.((((((....)))))).--------------)---)..)(((.(((....-----------------.)))))))))). ( -23.00) >consensus AAUGGUUUAAGCUAUCGAUGCGAGUGAACAUAUCCCAAGGGGAUGUG_________CCCC___UUUGGGGCAGGGGAGCCGUUCC______CCGGAUAACUCGGCAG ..................(((((((.....((((((...))))))...........((((......))))............................))))).)). (-12.26 = -12.93 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:02 2006