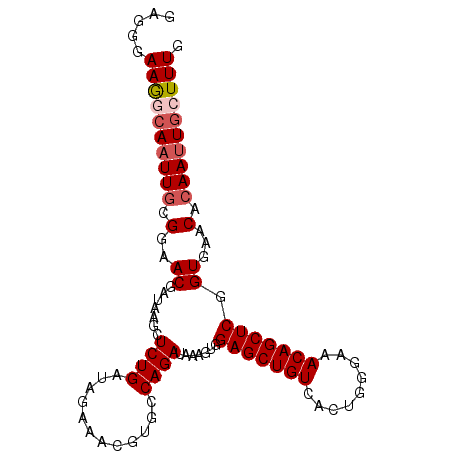
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
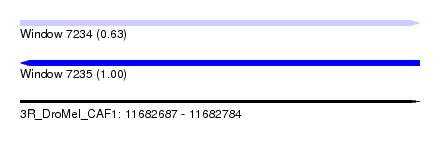
| Location | 11,682,687 – 11,682,784 |
| Length | 97 |
| Max. P | 0.999874 |

| Location | 11,682,687 – 11,682,784 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -20.78 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11682687 97 + 27905053 GAGGAAAGGCAAUUGCGGAACGAUAAGCUCUGAUAGAAACGUGCCAGAUAAAGUGGAGCUGUCACUGGGAAACAGCUCGGUGAACACAAUUGCUUUG .....((((((((((.(........(((((((.......(......)......))))))).((((((((......)))))))).).)))))))))). ( -28.62) >DroSec_CAF1 19294 97 + 1 GAGGGAAGGCAAUUGCGGAACGAUAAGCUCUGAUAGAAACGUGCCAGAUAAAGUGGAGCUGUCACUGGGAAACAGCUCGGUGAACACAAUUGCUUUG .....((((((((((.(........(((((((.......(......)......))))))).((((((((......)))))))).).)))))))))). ( -28.72) >DroSim_CAF1 22103 97 + 1 GAGGGAAGGCAAUUUCGGAACGAUAAGCUCUGAUAGAAACGUGCCAGAUAAAGUGGAGCUGUCACUGGGAAACAGCUCGGUGAACACAAUUGCUUUG .....((((((((((((((.(.....).))))).......((((((.......))).....((((((((......)))))))).)))))))))))). ( -25.80) >DroEre_CAF1 19382 91 + 1 A----AAAACAAUUGCGAAACGAUAAGCUCUGAUAGAAACGUGCCAGAUAAAGAGGAGCUGUGACUG--AAACAGCUCGGUGAACACAAUUACUUUG .----.....(((((.(..((......((((....................))))(((((((.....--..))))))).))...).)))))...... ( -17.05) >DroYak_CAF1 19450 95 + 1 GUGGGAAAACAAUUGCGGAACGAUAAGCUCUGAUAGAAACGUGCCAGAUAAAGUGGAGCUGUGACUG--UAACAGCUCGGUGAACACAACUGCUUUG .(((.(.........((((.(.....).)))).........).)))..((((((.(((((((.....--..)))))))(((.......))))))))) ( -21.47) >consensus GAGGGAAGGCAAUUGCGGAACGAUAAGCUCUGAUAGAAACGUGCCAGAUAAAGUGGAGCUGUCACUGGGAAACAGCUCGGUGAACACAAUUGCUUUG .....((((((((((.(..((.......((((............)))).......(((((((.........))))))).))...).)))))))))). (-20.78 = -21.54 + 0.76)


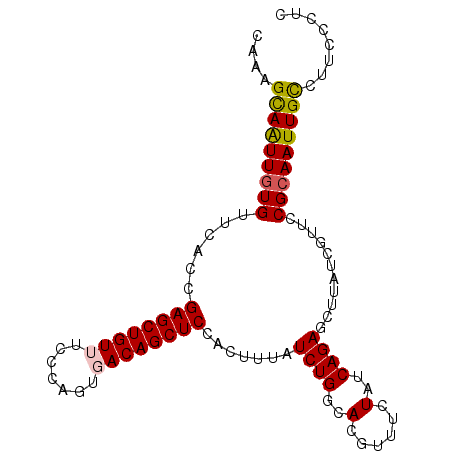
| Location | 11,682,687 – 11,682,784 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -22.28 |
| Energy contribution | -22.32 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.34 |
| SVM RNA-class probability | 0.999874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11682687 97 - 27905053 CAAAGCAAUUGUGUUCACCGAGCUGUUUCCCAGUGACAGCUCCACUUUAUCUGGCACGUUUCUAUCAGAGCUUAUCGUUCCGCAAUUGCCUUUCCUC ....(((((((((......((((((((.......)))))))).......((((..(......)..))))...........)))))))))........ ( -25.90) >DroSec_CAF1 19294 97 - 1 CAAAGCAAUUGUGUUCACCGAGCUGUUUCCCAGUGACAGCUCCACUUUAUCUGGCACGUUUCUAUCAGAGCUUAUCGUUCCGCAAUUGCCUUCCCUC ....(((((((((......((((((((.......)))))))).......((((..(......)..))))...........)))))))))........ ( -25.90) >DroSim_CAF1 22103 97 - 1 CAAAGCAAUUGUGUUCACCGAGCUGUUUCCCAGUGACAGCUCCACUUUAUCUGGCACGUUUCUAUCAGAGCUUAUCGUUCCGAAAUUGCCUUCCCUC ....((((((((((.....((((((((.......))))))))((.......)))))).......((.((((.....)))).))))))))........ ( -22.70) >DroEre_CAF1 19382 91 - 1 CAAAGUAAUUGUGUUCACCGAGCUGUUU--CAGUCACAGCUCCUCUUUAUCUGGCACGUUUCUAUCAGAGCUUAUCGUUUCGCAAUUGUUUU----U .((((((((((((..(...(((((((..--.....))))))).......((((..(......)..)))).......)...))))))))))))----. ( -19.60) >DroYak_CAF1 19450 95 - 1 CAAAGCAGUUGUGUUCACCGAGCUGUUA--CAGUCACAGCUCCACUUUAUCUGGCACGUUUCUAUCAGAGCUUAUCGUUCCGCAAUUGUUUUCCCAC .((((((((((((......(((((((..--.....))))))).......((((..(......)..))))...........))))))))))))..... ( -24.30) >consensus CAAAGCAAUUGUGUUCACCGAGCUGUUUCCCAGUGACAGCUCCACUUUAUCUGGCACGUUUCUAUCAGAGCUUAUCGUUCCGCAAUUGCCUUCCCUC ....(((((((((......((((((((.......)))))))).......((((..(......)..))))...........)))))))))........ (-22.28 = -22.32 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:58 2006