
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,651,760 – 11,651,960 |
| Length | 200 |
| Max. P | 0.959059 |

| Location | 11,651,760 – 11,651,880 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -30.84 |
| Energy contribution | -30.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11651760 120 + 27905053 CGAAAGUGCCAAGAGUCGAAGACAAUCAAUUAGAGGGUGCAUUACAAUCGGCGAUUCGAUGCAUCCCAUGACGAUGAUGAUGCUUCACAUCGUGAUUUUAGCAGCCACUUUUACGGCUGC (....).....((((((...............(.(((((((((..((((...)))).))))))))))...((((((.(((....))))))))))))))).((((((........)))))) ( -36.30) >DroSec_CAF1 32534 120 + 1 CGAAAGUGCCAAGAGUCGAAGACAAUCAAUUAGAGGGUGCAUUAUGAUCGGCCAUUCGAUGCAUCCCAUGACGACGAUGAUGCUUCACAUCGUGAUUUUAGCAGCCACUUUUACGGCUGC (....).....((((((............(((..((((((((((((......)))..)))))))))..)))..((((((........)))))))))))).((((((........)))))) ( -34.40) >DroSim_CAF1 34739 120 + 1 CGAAAGUGCCAAGAGUCGAAGACAAUCAAUUAGAGGGUGCAUUACGAUCGGCGAUUCGAUGCAUCCCAUGACGACGAUGAUGCUUCACAUCGUGAUUUUAGCAGCCACUUUUACGGCUGC (....).....((((((............(((..((((((((..(((........)))))))))))..)))..((((((........)))))))))))).((((((........)))))) ( -34.60) >DroEre_CAF1 30565 114 + 1 CGAAAGUGCCAAGAGUCGAAGACAAUCAAUUAGAGGGUGCAUUACGAUAGGCGAUUCGAUGCAUCCCAUG------AUGAUGCUUCACAUCGUGAUUUUAGCAGCCACUUUUACGGCUGC (....)...((.((((.((((...((((.(((..((((((((..(((........)))))))))))..))------))))).)))))).)).))......((((((........)))))) ( -34.80) >DroYak_CAF1 33246 114 + 1 CGAAAGUGCCAAGAGUCGAAGACAAUCAAUUAGAGGGUGCAUUACGAUAGGCGAUUCGAUGCAUCCCAUG------AUGAUGCUUCACAUCGUGAUUUUAGCAGCCACUUUUACGGCUGC (....)...((.((((.((((...((((.(((..((((((((..(((........)))))))))))..))------))))).)))))).)).))......((((((........)))))) ( -34.80) >consensus CGAAAGUGCCAAGAGUCGAAGACAAUCAAUUAGAGGGUGCAUUACGAUCGGCGAUUCGAUGCAUCCCAUGACGA_GAUGAUGCUUCACAUCGUGAUUUUAGCAGCCACUUUUACGGCUGC (....).....((((((...............(.((((((((..(((........)))))))))))).........((((((.....)))))))))))).((((((........)))))) (-30.84 = -30.88 + 0.04)



| Location | 11,651,760 – 11,651,880 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -29.96 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11651760 120 - 27905053 GCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCAUCAUCAUCGUCAUGGGAUGCAUCGAAUCGCCGAUUGUAAUGCACCCUCUAAUUGAUUGUCUUCGACUCUUGGCACUUUCG (((((((......))))))).........(((.(((.(((((.((((....))))))))).))).)))(((((..((.....((..((.....))..)).....))..)))))....... ( -34.00) >DroSec_CAF1 32534 120 - 1 GCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCAUCAUCGUCGUCAUGGGAUGCAUCGAAUGGCCGAUCAUAAUGCACCCUCUAAUUGAUUGUCUUCGACUCUUGGCACUUUCG (((((((......)))))))........(((.(((..(((((.((((....))))))))).(((((.(((.(((((................))))))))))))).......)))..))) ( -33.49) >DroSim_CAF1 34739 120 - 1 GCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCAUCAUCGUCGUCAUGGGAUGCAUCGAAUCGCCGAUCGUAAUGCACCCUCUAAUUGAUUGUCUUCGACUCUUGGCACUUUCG (((((((......))))))).......((((((........)))))).((((.(((.((((((((........)))..)))))))).....((((......))))....))))....... ( -33.90) >DroEre_CAF1 30565 114 - 1 GCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCAUCAU------CAUGGGAUGCAUCGAAUCGCCUAUCGUAAUGCACCCUCUAAUUGAUUGUCUUCGACUCUUGGCACUUUCG (((((((......))))))).....(((.((..((((((((((.------...(((.((((((((........)))..))))))))......))).)).)))))..)).)))........ ( -32.30) >DroYak_CAF1 33246 114 - 1 GCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCAUCAU------CAUGGGAUGCAUCGAAUCGCCUAUCGUAAUGCACCCUCUAAUUGAUUGUCUUCGACUCUUGGCACUUUCG (((((((......))))))).....(((.((..((((((((((.------...(((.((((((((........)))..))))))))......))).)).)))))..)).)))........ ( -32.30) >consensus GCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCAUCAUC_UCGUCAUGGGAUGCAUCGAAUCGCCGAUCGUAAUGCACCCUCUAAUUGAUUGUCUUCGACUCUUGGCACUUUCG (((((((......)))))))...(((((.((((.....))))...........(((.((((((((........)))..))))))))......)))))(((..........)))....... (-29.96 = -30.00 + 0.04)

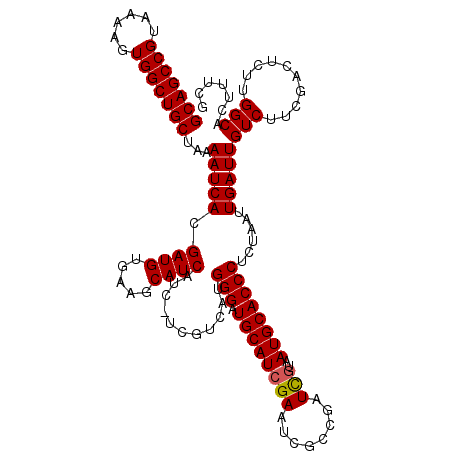

| Location | 11,651,800 – 11,651,920 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.63 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -28.00 |
| Energy contribution | -27.64 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11651800 120 + 27905053 AUUACAAUCGGCGAUUCGAUGCAUCCCAUGACGAUGAUGAUGCUUCACAUCGUGAUUUUAGCAGCCACUUUUACGGCUGCGCAGUGUUUUCAAUUGAACAGUAAGUGUGUAUUACAAUCA ............((((.((((((..(.((.((((((.(((....))))))))).))....((((((........))))))..(.(((((......))))).)..)..))))))..)))). ( -32.10) >DroSec_CAF1 32574 120 + 1 AUUAUGAUCGGCCAUUCGAUGCAUCCCAUGACGACGAUGAUGCUUCACAUCGUGAUUUUAGCAGCCACUUUUACGGCUGCGCAGUGCUUUCAAUUGAACAGUAAGUGUGUAUUACAAUCA ...((((((((....))))).))).........((((((........))))))((((.((((((((........))))))(((.(((((((....))).))))..)))....)).)))). ( -28.60) >DroSim_CAF1 34779 120 + 1 AUUACGAUCGGCGAUUCGAUGCAUCCCAUGACGACGAUGAUGCUUCACAUCGUGAUUUUAGCAGCCACUUUUACGGCUGCGCAGUGCUUUCAAUUGAACAGUAAGUGUGUAUUACAAUCA ..((((((((.((((..((.(((((.............))))).))..)))))))))...((((((........))))))(((.(((((((....))).))))..))))))......... ( -30.22) >DroEre_CAF1 30605 114 + 1 AUUACGAUAGGCGAUUCGAUGCAUCCCAUG------AUGAUGCUUCACAUCGUGAUUUUAGCAGCCACUUUUACGGCUGCGCAGUGCUUUCAAUUGAACAGUAAGUAUGCAUUACAAUCA ..........(((((..((.(((((.....------..))))).))..)))))((((.((((((((........))))))(((.(((((((....))).))))....)))..)).)))). ( -32.70) >DroYak_CAF1 33286 114 + 1 AUUACGAUAGGCGAUUCGAUGCAUCCCAUG------AUGAUGCUUCACAUCGUGAUUUUAGCAGCCACUUUUACGGCUGCGCAGUGCUUUCAAUUGAACAGUAAGUAUGCAUUGCAAUCA ..........(((((..((.(((((.....------..))))).))..)))))((((...((((((........))))))(((((((.....................))))))))))). ( -35.00) >consensus AUUACGAUCGGCGAUUCGAUGCAUCCCAUGACGA_GAUGAUGCUUCACAUCGUGAUUUUAGCAGCCACUUUUACGGCUGCGCAGUGCUUUCAAUUGAACAGUAAGUGUGUAUUACAAUCA ..........(((((..((.(((((.............))))).))..)))))((((.((((((((........))))))(((.(((((((....))).))))....)))..)).)))). (-28.00 = -27.64 + -0.36)


| Location | 11,651,800 – 11,651,920 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.63 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.24 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11651800 120 - 27905053 UGAUUGUAAUACACACUUACUGUUCAAUUGAAAACACUGCGCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCAUCAUCAUCGUCAUGGGAUGCAUCGAAUCGCCGAUUGUAAU (((..((((.......))))...)))...........((((((((((......)))))))...((((.((((.(((.(((((.((((....))))))))).))).))))..))))))).. ( -31.80) >DroSec_CAF1 32574 120 - 1 UGAUUGUAAUACACACUUACUGUUCAAUUGAAAGCACUGCGCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCAUCAUCGUCGUCAUGGGAUGCAUCGAAUGGCCGAUCAUAAU (((((((((.......)))).(..((.((((..(((((..(((((((......))))))).......(((((((((....))).))))))....)).))).)))).))..)))))).... ( -29.90) >DroSim_CAF1 34779 120 - 1 UGAUUGUAAUACACACUUACUGUUCAAUUGAAAGCACUGCGCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCAUCAUCGUCGUCAUGGGAUGCAUCGAAUCGCCGAUCGUAAU .(((((................(((....))).((.....(((((((......))))))).........(((.(((.(((((.((((....))))))))).))).))))))))))..... ( -30.80) >DroEre_CAF1 30605 114 - 1 UGAUUGUAAUGCAUACUUACUGUUCAAUUGAAAGCACUGCGCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCAUCAU------CAUGGGAUGCAUCGAAUCGCCUAUCGUAAU ((((((((.(((...(.....)(((....))).))).)))(((((((......)))))))...)))))((((.(((.(((((..------.....))))).))).))))........... ( -30.70) >DroYak_CAF1 33286 114 - 1 UGAUUGCAAUGCAUACUUACUGUUCAAUUGAAAGCACUGCGCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCAUCAU------CAUGGGAUGCAUCGAAUCGCCUAUCGUAAU ((((((((.(((...(.....)(((....))).))).)))(((((((......)))))))...)))))((((.(((.(((((..------.....))))).))).))))........... ( -32.70) >consensus UGAUUGUAAUACACACUUACUGUUCAAUUGAAAGCACUGCGCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCAUCAUC_UCGUCAUGGGAUGCAUCGAAUCGCCGAUCGUAAU ((((((..............((((........))))..(((((((((......))))))).........(((.(((.(((((.............))))).))).))))))))))).... (-25.12 = -25.24 + 0.12)


| Location | 11,651,840 – 11,651,960 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -21.38 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11651840 120 - 27905053 CUCAUAAACAAAAUUAACACGGAACAUAAAUCUUUCCCAUUGAUUGUAAUACACACUUACUGUUCAAUUGAAAACACUGCGCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCA .(((................((((.........)))).(((((..((((.......))))...)))))))).......(((((((((......))))))).....((((...)))).)). ( -21.90) >DroSec_CAF1 32614 120 - 1 CUCAUAAACAAAAUUAACACGGAACAUAAAUCUUUCCCAUUGAUUGUAAUACACACUUACUGUUCAAUUGAAAGCACUGCGCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCA ...............................(((((..(((((..((((.......))))...))))).)))))....(((((((((......))))))).....((((...)))).)). ( -23.70) >DroSim_CAF1 34819 120 - 1 CUCAUAAACAAAAUUAACACGGAACAUAAAUCUUUCCCAUUGAUUGUAAUACACACUUACUGUUCAAUUGAAAGCACUGCGCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCA ...............................(((((..(((((..((((.......))))...))))).)))))....(((((((((......))))))).....((((...)))).)). ( -23.70) >DroEre_CAF1 30639 120 - 1 CUCAUAAACAAAAUUAACACGGACCAUAAAUCUUUCCCACUGAUUGUAAUGCAUACUUACUGUUCAAUUGAAAGCACUGCGCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCA .(((((..............(((...........)))...((((((((.(((...(.....)(((....))).))).)))(((((((......)))))))...)))))...))))).... ( -22.20) >DroYak_CAF1 33320 120 - 1 CUCAUAAACAAAAUUAACACGGACAUUAAAUCUUUCCCAUUGAUUGCAAUGCAUACUUACUGUUCAAUUGAAAGCACUGCGCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCA ................................((((.(((((...(((.(((...(.....)(((....))).))).)))(((((((......)))))))........))))).)))).. ( -24.40) >consensus CUCAUAAACAAAAUUAACACGGAACAUAAAUCUUUCCCAUUGAUUGUAAUACACACUUACUGUUCAAUUGAAAGCACUGCGCAGCCGUAAAAGUGGCUGCUAAAAUCACGAUGUGAAGCA ...............................(((((..(((((..((((.......))))...))))).)))))....(((((((((......))))))).....((((...)))).)). (-21.38 = -21.98 + 0.60)

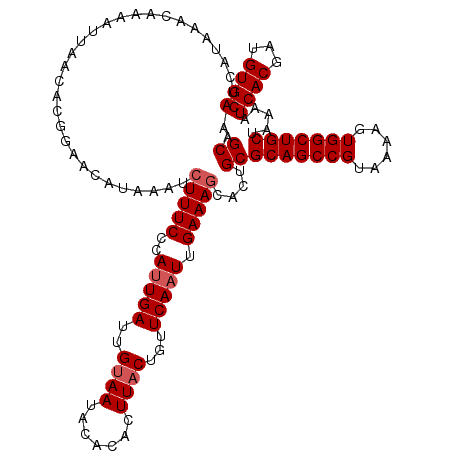
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:44 2006