
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,648,139 – 11,648,539 |
| Length | 400 |
| Max. P | 0.999865 |

| Location | 11,648,139 – 11,648,259 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -40.76 |
| Consensus MFE | -36.66 |
| Energy contribution | -37.02 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11648139 120 + 27905053 CCCGGGUGGUCAGCAUGUUACGCAGCCACCGAGUUACAAGGUUCUCAGUCAGCAGGCUCAGCAGCUGGCAAAUGCCUUGCGUUACCCGCCGAAUCUGUACUUUGUAAGCAACCUGGAGCA .(((((((((..((.((.....)))).)))...(((((((((...(((((.((.((....((((..(((....)))))))....)).)).))..))).)))))))))...)))))).... ( -38.70) >DroSec_CAF1 28980 120 + 1 CCCGGGUGGCCAGCAUGUGACACAGCCACCGAGCUACAAGGUACUCAGUCAGCAGGCGCAGCAGCUGGCAAAUGCUUUACGUUACCCGCCGAAUCUGUACUUUGUAAGCAACCUGGAGCA .((((((((...((.((.....))))..))..((((((((((((....((.((.((.(.(((....(((....)))....))).))))).))....))))))))).))).)))))).... ( -37.90) >DroSim_CAF1 31180 120 + 1 CCCGGCUGGUCAGCAUGUGACACAGCCACCGAGCUACAAGGUACUCAGUCAGCAGGCGCAGCAGCUGGCAAAUGCCUUACGUUACCCGCCGAAUCUGUACUUUGUAAGCAACCUGGAGCA .(((((((((((.....)))).))))).....((((((((((((....((.((.((.(.(((....(((....)))....))).))))).))....))))))))).))).....)).... ( -42.90) >DroEre_CAF1 26884 120 + 1 CCCGGGUGGUCAGCAUGUGACUCAGCCACCGAGCUACAAGGUACUCAGUCAGCAGGCUCAGCACCUGGCAAAUGCCUUGCGUUACCCGCCGAAUCUGUACUUUGUAAGCAAUCUGGAGCA .(((((((((.(((....).))..))))))..((((((((((((....((.((.((....(((...(((....))).)))....)).)).))....))))))))).)))....))).... ( -41.30) >DroYak_CAF1 29482 120 + 1 CCCGGGUGGCCAGCAUGUGACUCAGCCACCGAGCUACAAGGUACUCAGUCAGCAGGCUCAGCACCUGGCAAAUGCCUUGCGUUACCCGCCGAAUCUGUACUUUGUAAGCAACCUGGAGCA .(((((((((.(((....).))..))).....((((((((((((....((.((.((....(((...(((....))).)))....)).)).))....))))))))).))).)))))).... ( -43.00) >consensus CCCGGGUGGUCAGCAUGUGACACAGCCACCGAGCUACAAGGUACUCAGUCAGCAGGCUCAGCAGCUGGCAAAUGCCUUGCGUUACCCGCCGAAUCUGUACUUUGUAAGCAACCUGGAGCA .((((((((...((.((.....))))..))..((((((((((((....((.((.((....(((...(((....))).)))....)).)).))....))))))))).))).)))))).... (-36.66 = -37.02 + 0.36)

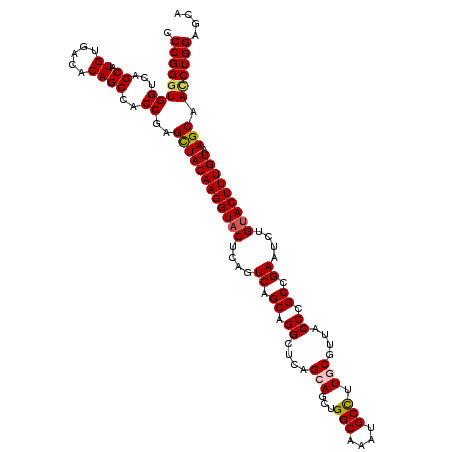
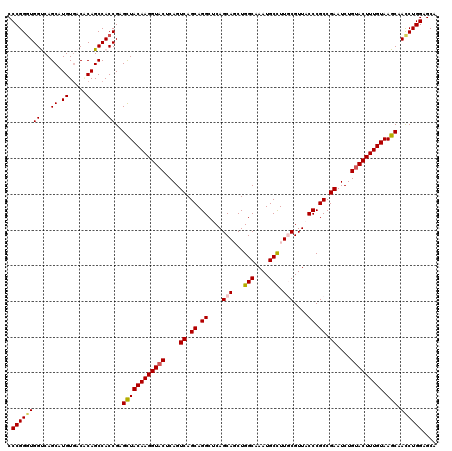
| Location | 11,648,179 – 11,648,299 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -46.58 |
| Consensus MFE | -40.22 |
| Energy contribution | -40.42 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11648179 120 - 27905053 AAGCCUGACCAAAGGCAGCAAUGCAGCGGCCAGUUCUGGCUGCUCCAGGUUGCUUACAAAGUACAGAUUCGGCGGGUAACGCAAGGCAUUUGCCAGCUGCUGAGCCUGCUGACUGAGAAC ..((((......))))(((((((.((((((((....)))))))).))..))))).........(((..(((((((((...(((.(((....)))...)))...))))))))))))..... ( -49.10) >DroSec_CAF1 29020 120 - 1 AAGCCUGACCAAAGGCAGCAAGGCAGAGGCCAGUUCUGGCUGCUCCAGGUUGCUUACAAAGUACAGAUUCGGCGGGUAACGUAAAGCAUUUGCCAGCUGCUGCGCCUGCUGACUGAGUAC ..((((......)))).((..(((((((((((((.(((((((((..(.((((((..(..(((....)))..)..)))))).)..))))...)))))..)))).)))).))).))..)).. ( -45.60) >DroSim_CAF1 31220 120 - 1 AAGCCUGACCAAAGGCAGCAAUGAAGCGGCCAGUUCUGGCUGCUCCAGGUUGCUUACAAAGUACAGAUUCGGCGGGUAACGUAAGGCAUUUGCCAGCUGCUGCGCCUGCUGACUGAGUAC ..((((......))))(((((((.((((((((....)))))))).))..)))))......((((....(((((((((.......(((....))).((....)))))))))))....)))) ( -49.60) >DroEre_CAF1 26924 120 - 1 AAGCCUGACCAAAGGCAGCAAUGCAGAGGCCAGUUCUGGCUGCUCCAGAUUGCUUACAAAGUACAGAUUCGGCGGGUAACGCAAGGCAUUUGCCAGGUGCUGAGCCUGCUGACUGAGUAC ..((((......))))(((((((.((.(((((....))))).)).))..)))))......((((....(((((((((...(((.(((....)))...)))...)))))))))....)))) ( -44.70) >DroYak_CAF1 29522 120 - 1 AAGCCUGACCAAAGGCAGAAGUGCAGAGGCCAGUUCUGGCUGCUCCAGGUUGCUUACAAAGUACAGAUUCGGCGGGUAACGCAAGGCAUUUGCCAGGUGCUGAGCCUGCUGACUGAGUAC ..((((......))))...(((.(((((((((((((((((((((....((((((..(..(((....)))..)..))))))....))))...)))))).)))).)))).))))))...... ( -43.90) >consensus AAGCCUGACCAAAGGCAGCAAUGCAGAGGCCAGUUCUGGCUGCUCCAGGUUGCUUACAAAGUACAGAUUCGGCGGGUAACGCAAGGCAUUUGCCAGCUGCUGAGCCUGCUGACUGAGUAC ..((((......))))(((((((.((.(((((....))))).)).))..))))).........(((..(((((((((...(((.(((....)))...)))...))))))))))))..... (-40.22 = -40.42 + 0.20)

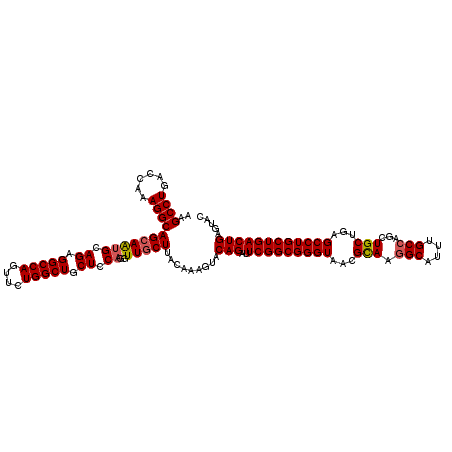
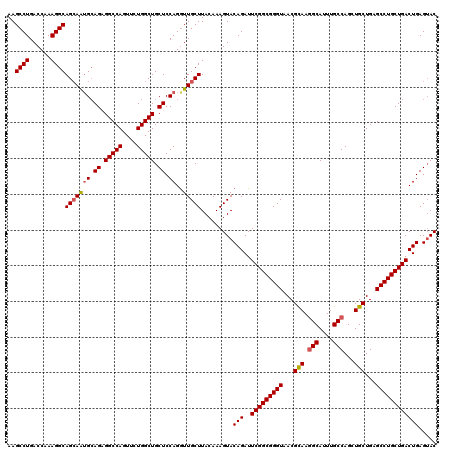
| Location | 11,648,219 – 11,648,339 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -43.74 |
| Consensus MFE | -38.38 |
| Energy contribution | -38.46 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11648219 120 - 27905053 UGGUUAGGUGUACGUCACAGGGCAGCGGUCCCUUAGGCAAAAGCCUGACCAAAGGCAGCAAUGCAGCGGCCAGUUCUGGCUGCUCCAGGUUGCUUACAAAGUACAGAUUCGGCGGGUAAC (((((((((....(((..(((((....).))))..)))....))))))))).(((((((..((.((((((((....)))))))).)).)))))))......................... ( -45.40) >DroSec_CAF1 29060 120 - 1 UGGUCAGGUGUACGUCACAGGGCAGCGGUCCCUUAGGCAAAAGCCUGACCAAAGGCAGCAAGGCAGAGGCCAGUUCUGGCUGCUCCAGGUUGCUUACAAAGUACAGAUUCGGCGGGUAAC (((((((((....(((..(((((....).))))..)))....))))))))).(((((((..((.((.(((((....))))).))))..)))))))......................... ( -44.80) >DroSim_CAF1 31260 120 - 1 UGGUCAGGUGUACGUCACAGGGCAGCGGUCCUUUAGGCAAAAGCCUGACCAAAGGCAGCAAUGAAGCGGCCAGUUCUGGCUGCUCCAGGUUGCUUACAAAGUACAGAUUCGGCGGGUAAC (((((((((....(((..(((((....)))))...)))....))))))))).(((((((..((.((((((((....)))))))).)).)))))))......................... ( -48.30) >DroEre_CAF1 26964 120 - 1 UGGUCAGGUGCACGUCACAGGGCAGCGGUCCCUUAGGCAAAAGCCUGACCAAAGGCAGCAAUGCAGAGGCCAGUUCUGGCUGCUCCAGAUUGCUUACAAAGUACAGAUUCGGCGGGUAAC (((((((((....(((..(((((....).))))..)))....)))))))))...(((....)))((.(((((....))))).)).....(((((..(..(((....)))..)..))))). ( -39.60) >DroYak_CAF1 29562 120 - 1 UGGUCAGGUGCACGUCGCAGGGCAGCGGUCCCUUAGGCAAAAGCCUGACCAAAGGCAGAAGUGCAGAGGCCAGUUCUGGCUGCUCCAGGUUGCUUACAAAGUACAGAUUCGGCGGGUAAC (((((((((....(((..(((((....).))))..)))....))))))))).((.(((((.(((....).)).))))).)).......((((((..(..(((....)))..)..)))))) ( -40.60) >consensus UGGUCAGGUGUACGUCACAGGGCAGCGGUCCCUUAGGCAAAAGCCUGACCAAAGGCAGCAAUGCAGAGGCCAGUUCUGGCUGCUCCAGGUUGCUUACAAAGUACAGAUUCGGCGGGUAAC (((((((((....(((..(((((....).))))..)))....))))))))).(((((((...(.((.(((((....))))).)).)..)))))))......................... (-38.38 = -38.46 + 0.08)


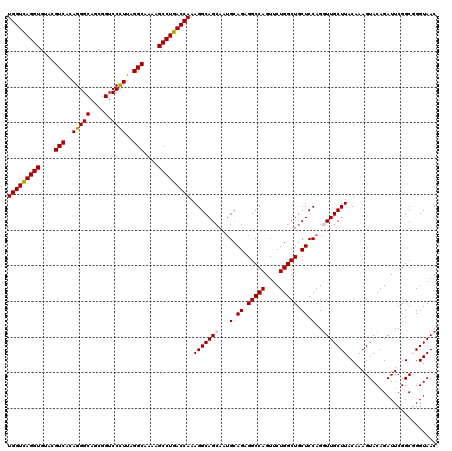
| Location | 11,648,339 – 11,648,459 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -41.72 |
| Consensus MFE | -39.40 |
| Energy contribution | -40.40 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11648339 120 - 27905053 CCUCCAUAGACAAUUCCCUGCCAGCGCUUACACUGCAAUCGAAUCCUUGGCGAUGCAAAACUAGCAGCGCCGAGGCGGACGGAAACAGUUGCAACUCCGGAUCGGAUAGGGUACGCAGGG ...............((((((..((.((((...((((((((...((((((((.(((.......))).))))))))))...(....).))))))..((((...))))))))))..)))))) ( -41.00) >DroSec_CAF1 29180 120 - 1 CCUCCAUAGACAAUUCCCUGCCAGUGCUCACACUGCAAUCGAAUCCUUGACGAUGUAAAACUAGCAGCGCCGAGGCGGACGGAAACAGUUGCAGCUCCGGAUCGGAUAGGGUACGCAGGG ...............((((((..((((((...(((((((((...(((((.((.(((.......))).)).)))))))...(....).))))))).((((...))))..)))))))))))) ( -39.50) >DroSim_CAF1 31380 120 - 1 ACUCCAUAGACAAUUCCCUGCCAGCGCUCACACUGCAAUCGAAUCCUUGGCGAUGCAAAACUAGCAGCGCCGAGGCGGACGGAAACAGUUGCAGCUCCGGAUCUGAUAGGGUACGCAGGG ...............((((((..(..(((...(((((((((...((((((((.(((.......))).))))))))))...(....).)))))))...((....))...)))..))))))) ( -40.40) >DroEre_CAF1 27084 120 - 1 AAUCCAGAGACAAUUCACUGCCAGCGCUCACACUGCAAUCGAAUCCUUGGCGAUGCAAAACUAGCAGCGCCGAGGCGGACGGAAACAGUUGCAGCUCCGGAUCGGAUAGGGUGCGCAGGG .................((((..((((((...(((((((((...((((((((.(((.......))).))))))))))...(....).))))))).((((...))))..)))))))))).. ( -45.20) >DroYak_CAF1 29682 120 - 1 AGUCCAUAGACAAUUCCCUGCCAGCGCUCACACUGCAAUCGAAUCCUUGGCGAUGCAAAACUAGCAGCGCCGAGGCGGACGGAAACAGUUGCAGCUCCGGAUCGGAUAAAGUGCGCAGCG .(((....)))......((((..(((((....(((((((((...((((((((.(((.......))).))))))))))...(....).))))))).((((...))))...))))))))).. ( -42.50) >consensus ACUCCAUAGACAAUUCCCUGCCAGCGCUCACACUGCAAUCGAAUCCUUGGCGAUGCAAAACUAGCAGCGCCGAGGCGGACGGAAACAGUUGCAGCUCCGGAUCGGAUAGGGUACGCAGGG ...............((((((..((((((...(((((((((...((((((((.(((.......))).))))))))))...(....).))))))).((((...))))..)))))))))))) (-39.40 = -40.40 + 1.00)

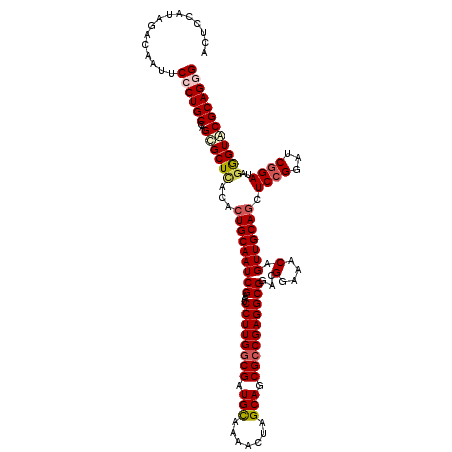

| Location | 11,648,379 – 11,648,499 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -38.98 |
| Energy contribution | -39.30 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.30 |
| SVM RNA-class probability | 0.999865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11648379 120 - 27905053 UGCACAUUACCCAAUCCCUUGCUGGACAGGGGACAUAUGGCCUCCAUAGACAAUUCCCUGCCAGCGCUUACACUGCAAUCGAAUCCUUGGCGAUGCAAAACUAGCAGCGCCGAGGCGGAC ..............(((...(((((.(((((((..(((((...))))).....))))))))))))((.......))........((((((((.(((.......))).)))))))).))). ( -45.20) >DroSec_CAF1 29220 120 - 1 UGCACAUUACCCAAACCCUUGCUGGACAGGGGACAUAUGGCCUCCAUAGACAAUUCCCUGCCAGUGCUCACACUGCAAUCGAAUCCUUGACGAUGUAAAACUAGCAGCGCCGAGGCGGAC ..........((........(((((.(((((((..(((((...))))).....))))))))))))((.......))........(((((.((.(((.......))).)).))))).)).. ( -33.30) >DroSim_CAF1 31420 120 - 1 UGCACAUUACCCAAUCCCUUGCUGGACAGGGGACAUAUGGACUCCAUAGACAAUUCCCUGCCAGCGCUCACACUGCAAUCGAAUCCUUGGCGAUGCAAAACUAGCAGCGCCGAGGCGGAC ..............(((...(((((.(((((((..(((((...))))).....))))))))))))((.......))........((((((((.(((.......))).)))))))).))). ( -45.20) >DroEre_CAF1 27124 120 - 1 UGUACAUUUCCCAAUCCCUUGCUGGACAGCGGACAUAUGGAAUCCAGAGACAAUUCACUGCCAGCGCUCACACUGCAAUCGAAUCCUUGGCGAUGCAAAACUAGCAGCGCCGAGGCGGAC ..............(((...(((((.(((.(((....(((...))).......))).))))))))((.......))........((((((((.(((.......))).)))))))).))). ( -36.20) >DroYak_CAF1 29722 120 - 1 UGCACAUUACCCAAUCCCUUGCUGGACAGAGGGCAUAUGGAGUCCAUAGACAAUUCCCUGCCAGCGCUCACACUGCAAUCGAAUCCUUGGCGAUGCAAAACUAGCAGCGCCGAGGCGGAC ..............(((...(((((.(((.(((..(((((...)))))......)))))))))))((.......))........((((((((.(((.......))).)))))))).))). ( -39.50) >consensus UGCACAUUACCCAAUCCCUUGCUGGACAGGGGACAUAUGGACUCCAUAGACAAUUCCCUGCCAGCGCUCACACUGCAAUCGAAUCCUUGGCGAUGCAAAACUAGCAGCGCCGAGGCGGAC ..........((........(((((.(((((((..(((((...))))).....))))))))))))((.......))........((((((((.(((.......))).)))))))).)).. (-38.98 = -39.30 + 0.32)

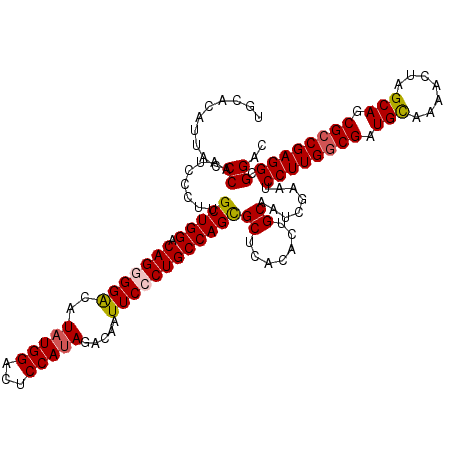
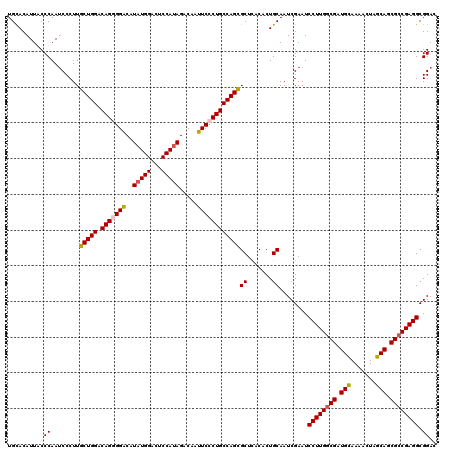
| Location | 11,648,419 – 11,648,539 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -34.96 |
| Energy contribution | -35.64 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11648419 120 - 27905053 GCUGGUGGCUUCGAUUGUAUGCAGGCUGAGCCGACCUAGCUGCACAUUACCCAAUCCCUUGCUGGACAGGGGACAUAUGGCCUCCAUAGACAAUUCCCUGCCAGCGCUUACACUGCAAUC (((((((((((.(.(((....))).).))))).))).)))....................(((((.(((((((..(((((...))))).....))))))))))))((.......)).... ( -41.40) >DroSec_CAF1 29260 120 - 1 GCUGGUGGCUUCGAUCGUAUGCAGGCUGAGCCGACCCAGCUGCACAUUACCCAAACCCUUGCUGGACAGGGGACAUAUGGCCUCCAUAGACAAUUCCCUGCCAGUGCUCACACUGCAAUC ((..(((((.......((((((((.(((........))))))))...)))..........(((((.(((((((..(((((...))))).....)))))))))))))).)))...)).... ( -39.90) >DroSim_CAF1 31460 120 - 1 GCUGGUGGCUUCGAUCGUAUGCAGGCUGAGCCGACCCAGCUGCACAUUACCCAAUCCCUUGCUGGACAGGGGACAUAUGGACUCCAUAGACAAUUCCCUGCCAGCGCUCACACUGCAAUC ((..(((((...(((.((((((((.(((........))))))))...)))...)))....(((((.(((((((..(((((...))))).....)))))))))))))).)))...)).... ( -42.50) >DroEre_CAF1 27164 120 - 1 ACUGGUGGCUUCGAUUGUAUGCAGGCUGAGCCGACCCAGCUGUACAUUUCCCAAUCCCUUGCUGGACAGCGGACAUAUGGAAUCCAGAGACAAUUCACUGCCAGCGCUCACACUGCAAUC ....(((((...(((((.(((..(((((........)))))...)))....)))))....(((((.(((.(((....(((...))).......))).)))))))))).)))......... ( -31.90) >DroYak_CAF1 29762 120 - 1 ACUGGUGGCUUCGAUUGUAUGCAGGCUCAGCCGACCCAGUUGCACAUUACCCAAUCCCUUGCUGGACAGAGGGCAUAUGGAGUCCAUAGACAAUUCCCUGCCAGCGCUCACACUGCAAUC ((((((((((....(((....)))....)))))..)))))....................(((((.(((.(((..(((((...)))))......)))))))))))((.......)).... ( -32.60) >consensus GCUGGUGGCUUCGAUUGUAUGCAGGCUGAGCCGACCCAGCUGCACAUUACCCAAUCCCUUGCUGGACAGGGGACAUAUGGACUCCAUAGACAAUUCCCUGCCAGCGCUCACACUGCAAUC (((((((((((.(.(((....))).).))))))..)))))....................(((((.(((((((..(((((...))))).....))))))))))))((.......)).... (-34.96 = -35.64 + 0.68)

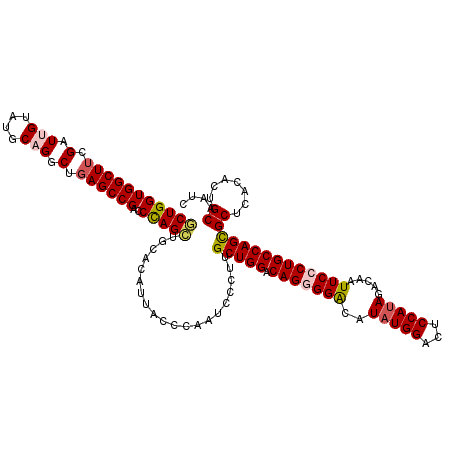
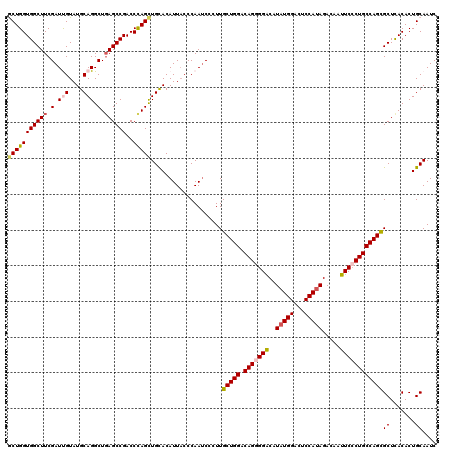
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:35 2006