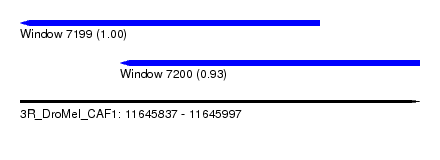
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,645,837 – 11,645,997 |
| Length | 160 |
| Max. P | 0.995611 |

| Location | 11,645,837 – 11,645,957 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -35.00 |
| Energy contribution | -34.92 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11645837 120 - 27905053 UACUCGGUGUACUCGCCUGGAAUUUUGCGAAAGUCAAAGUUCAGACAUAUGAAGGGAUGUGGGCCACACGAUCCCUUAAUCGAGUAUAUAUCGAAGGGCAUGUUGUGAGUGAGAAGGCUA (((((((((((((((.(((.((((((((....).)))))))))).......((((((((((.....))).)))))))...))))))))).((....)).......))))))......... ( -39.60) >DroSec_CAF1 26689 120 - 1 UACUCGGUGUACUCGCCUGGAAUUUUGCGGAAGUCGAAGUUUAAACAGAUGAAGGGAUGUGGGCCACACGAUCCCUUAAUCGAGUAUAUAUCGAAGGGCAUGUUGUGAGUGAGCAGGCUA ...(((((((((((.....(((((((((....).)))))))).....(((.((((((((((.....))).))))))).)))))))))..)))))..(((.((((.......)))).))). ( -39.90) >DroSim_CAF1 28875 120 - 1 UACUCGGUGUACUCGCCUGGAAUUUUGCGGAAAUCGAAGUUUAGACAGAUGAAGGGAUGUGGGCCACAUGAUCCCUUAAUCGAGUAUAUAUCGAAGGGCAUGUUGUGAGUGAGCAGGCUA ...((((((((((((.(((.(((((((.......)))))))))).).(((.((((((((((.....))).))))))).)))))))))..)))))..(((.((((.......)))).))). ( -33.70) >DroEre_CAF1 24583 120 - 1 UACUCGGUGUACUCGCCCGGAAUUUUGCGGAAGUCAAAGUUUAGACAGAUGAAAGGAUGUGGGCCACACGAUCCCUUAAUUGAGUAUAUAUCGAAGGGCAUGUUGUGAGUGAGUAGGCUA .....(((.(((((((((.(((((((((....).))))))))..(((..(....)..)))))))..((((((((((...((((.......))))))))...))))))...))))).))). ( -39.60) >DroYak_CAF1 27171 120 - 1 UACUCGGUGUACUCGCCCGGAAUUUUGCGGAAGUCAAAGUUUAGACAGAUGAAGGGAUGUGGGCCGCACGAUCCCUUAAUCGAGUAUAUAUCGAAGGGCAUGUUGUGAGUGAGCAGGCUA ...(((((((((((.....(((((((((....).)))))))).....(((.((((((((((.....))).))))))).)))))))))..)))))..(((.((((.......)))).))). ( -39.20) >consensus UACUCGGUGUACUCGCCUGGAAUUUUGCGGAAGUCAAAGUUUAGACAGAUGAAGGGAUGUGGGCCACACGAUCCCUUAAUCGAGUAUAUAUCGAAGGGCAUGUUGUGAGUGAGCAGGCUA ((((((((((((((.....(((((((((....).)))))))).....(((.((((((((((.....))).))))))).))))))))))).((....)).......))))))......... (-35.00 = -34.92 + -0.08)

| Location | 11,645,877 – 11,645,997 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -34.78 |
| Energy contribution | -35.54 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11645877 120 - 27905053 CUCCAGAUUGUCGUCUGUGAUGAACUGCGCCUUAACGGAGUACUCGGUGUACUCGCCUGGAAUUUUGCGAAAGUCAAAGUUCAGACAUAUGAAGGGAUGUGGGCCACACGAUCCCUUAAU ...((...((((......((.(.(((.((......)).))).)))((((....))))..(((((((((....).)))))))).))))..))((((((((((.....))).)))))))... ( -37.70) >DroSec_CAF1 26729 120 - 1 CUCCAGAUUGUCGUCUGUAAUGAACUGCGCCUUAACAGAGUACUCGGUGUACUCGCCUGGAAUUUUGCGGAAGUCGAAGUUUAAACAGAUGAAGGGAUGUGGGCCACACGAUCCCUUAAU ..........((((((((..((((((.((.(((..((((((.((.((((....)))).)).))))))...))).)).)))))).))))))))(((((((((.....))).)))))).... ( -40.60) >DroSim_CAF1 28915 120 - 1 CUCCAGAUUGUCGUCUGUAAUGAACUGCGCCUUAACAGAGUACUCGGUGUACUCGCCUGGAAUUUUGCGGAAAUCGAAGUUUAGACAGAUGAAGGGAUGUGGGCCACAUGAUCCCUUAAU ..........((((((((..((((((.((..((..((((((.((.((((....)))).)).))))))..))...)).)))))).))))))))(((((((((.....))).)))))).... ( -36.20) >DroEre_CAF1 24623 120 - 1 CUCUAGAUUGUCGUCUGUGAUGAACUGUGCCUUAACGGAGUACUCGGUGUACUCGCCCGGAAUUUUGCGGAAGUCAAAGUUUAGACAGAUGAAAGGAUGUGGGCCACACGAUCCCUUAAU ..........((((((((..((((((.((.(((....((((((.....))))))..(((........)))))).)).)))))).))))))))..(((((((.....))).))))...... ( -37.10) >DroYak_CAF1 27211 120 - 1 CUCUAGAUUGUCGUCAGUAAUGAACUGCGCCUUAACUGAGUACUCGGUGUACUCGCCCGGAAUUUUGCGGAAGUCAAAGUUUAGACAGAUGAAGGGAUGUGGGCCGCACGAUCCCUUAAU .......((((((.((((.....))))).........((((((.....)))))).....(((((((((....).)))))))).)))))...((((((((((.....))).)))))))... ( -36.00) >consensus CUCCAGAUUGUCGUCUGUAAUGAACUGCGCCUUAACAGAGUACUCGGUGUACUCGCCUGGAAUUUUGCGGAAGUCAAAGUUUAGACAGAUGAAGGGAUGUGGGCCACACGAUCCCUUAAU ..........((((((((..((((((.((.(((..((((((.((.((((....)))).)).))))))...))).)).)))))).))))))))(((((((((.....))).)))))).... (-34.78 = -35.54 + 0.76)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:23 2006