
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,633,966 – 11,634,078 |
| Length | 112 |
| Max. P | 0.509898 |

| Location | 11,633,966 – 11,634,078 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.09 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -16.01 |
| Energy contribution | -18.43 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.46 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11633966 112 - 27905053 ACAAAGGAGUCCGACCAGAACACUGGAGAAUGCUCCUCCAAGUAAUCCCAGUAACUUCCAGCCAUUUUAGUUGG---AAAAAAAUGGGGGGUGUUGUG--GGGCG---GAAGGUCGUUGU ((((.((..((((.(((.((((((((((.......))))......(((((.....(((((((.......)))))---)).....))))))))))).))--)..))---))...)).)))) ( -42.60) >DroSec_CAF1 15144 112 - 1 ACAAAGGAGCCCGACCAGAACACUGGAGAAUGCUCCUCCAAGUAAUCCCAGUAACUUCCAGCCAUUUUAGUUGG---AAAAAAAUGGGGGGUGUUGUG--GGGCG---GAAGGUCGUUGU ((((..((.((((.(((.((((((((((.......))))......(((((.....(((((((.......)))))---)).....))))))))))).))--)..))---)..).)).)))) ( -40.30) >DroSim_CAF1 12439 115 - 1 ACAAAGGAGCCCGACCAGAACACUGAAGAAUGCUCCUCCAAAUAAUCCCAGUAACUUCCAGCCAUUUUAGUUGGGAAAAAAAAAUGGGGGGUGUUGUG--GGGCG---GAAGGUCGUUGU ((((..((.((((.(((.((((((((.((....)).)).......(((((.....(((((((.......)))))))........))))))))))).))--)..))---)..).)).)))) ( -32.32) >DroEre_CAF1 12791 106 - 1 ACAAAGGAACCCGACCAGAACACUGGAGAAUACUCCUCCA-----GCCCAGUAACUUCCAGCCGUUUUAGUUGG---AA-AAAAUGGGGGGUGUUGUG--GGGCA---GAAGGUCGUUGU ...........(((((.......(((((.......)))))-----((((..((((.(((..(((((((......---..-))))))).))).))))..--)))).---...))))).... ( -36.60) >DroYak_CAF1 15247 111 - 1 ACAAAGGAACCCGACCAGAACACUGGAGAAUGCUCCUCCAAGUAAUCCCAGUGACUUCCAGCCAUUUUACUUGG---CC-CAAAUGGGGGGUGUUGUG--GGGCA---GAAGGUCGUUGU ...........(((((....((((((.((.((((......)))).)))))))).((.((..((((..(((((..---((-(....))))))))..)))--))).)---)..))))).... ( -36.90) >DroAna_CAF1 14406 96 - 1 ACAAAGGAAUCC--CGAAUCCGCUGGAGA----------------GCCCAGUAACUUUGAGGC---UCAAACGCG--AA-ACAAAGUGAAAAGCCGGGAGAGGAAAAUAAAGGUCGUUGU .........(((--((.....(((...((----------------((((((.....))).)))---))...(((.--..-.....)))...))))))))..................... ( -22.30) >consensus ACAAAGGAACCCGACCAGAACACUGGAGAAUGCUCCUCCAAGUAAUCCCAGUAACUUCCAGCCAUUUUAGUUGG___AA_AAAAUGGGGGGUGUUGUG__GGGCA___GAAGGUCGUUGU ...........(((((.......(((((.......)))))......(((.(((((.(((..(((((((............))))))).))).)))))...)))........))))).... (-16.01 = -18.43 + 2.42)

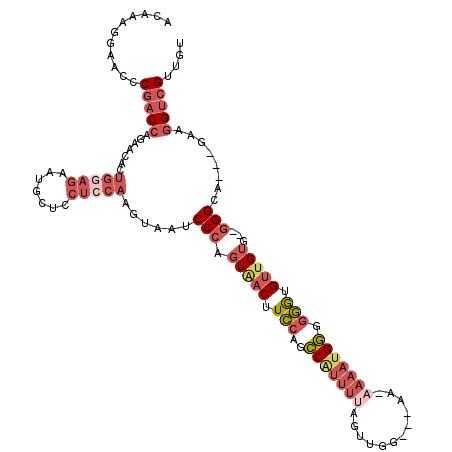
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:09 2006