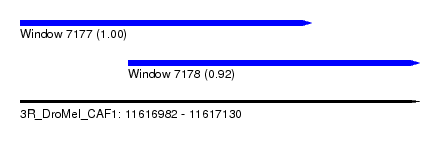
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,616,982 – 11,617,130 |
| Length | 148 |
| Max. P | 0.995670 |

| Location | 11,616,982 – 11,617,090 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.54 |
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -29.30 |
| Energy contribution | -30.50 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11616982 108 + 27905053 CUAUUGGCAGCUGCUCAAUGCGAUCUUCAAGCUGGGCGCAGUCAAGCAUCUUUGCAUUUAAGUCGCUGCUUAAUUGUAAAAUGUUUGCCUGUGG---AGAACAUAAUGU---------UA .(((((.(((((((.....))(.....).)))))..(((((.(((((((.((((((.((((((....)))))).))))))))))))).))))).---......))))).---------.. ( -31.20) >DroSec_CAF1 6918 120 + 1 CUAUUGGUAGCUGCUCAAUGCGGUCUUCAAGCUCGGCGCAGGUAAGCAACUUUGCAUUUAAGUCGCUGCUUAAUUGUAAAAUGCUUGCCUGUGGAGGAGAACAUAAUGUCAAUAAUCUUA ...((((..(((((.....)))))..)))).(((..((((((((((((..((((((.((((((....)))))).)))))).))))))))))))...)))..................... ( -41.20) >DroSim_CAF1 7046 120 + 1 CUAAUGGCAGCUGCUCAAUGCGAACUUCAAGCUCGGCGCAGGCAAGCAUCUUUGCAUUUAAGUCGCUGCUUAAUUGUAAAAUGUUUGCCUGUGGAGGAGAACAUAAUAUUAUUAAUCAUA ...(((....((.(((....(((.(.....).))).(((((((((((((.((((((.((((((....)))))).)))))))))))))))))))))).))..)))................ ( -39.90) >DroEre_CAF1 4510 117 + 1 CUAUUGGCAUCUGCUCAAUGCGCUCUGCAAGCUCGGUGCAGGCAAGAAUCAUUGCAUUUAAGUCGGUGCUUAGUUGUAAGAUGUUUGCCUGUGG---AGAAAAUAAUGUUAGUAAUCCUA .(((((((((..((.....)).(((..(..((.....))(((((((.(((.(((((.((((((....)))))).)))))))).))))))))..)---))......)))))))))...... ( -36.30) >DroYak_CAF1 5006 117 + 1 CUAUUGUCACCUGCUCAUUGCGCUCUGCAAGCUCGUCGCAGGCUUGCAUCUUUGCAUUUAAGUCGGAGCUUAGUUGUAAGAUGUUUGCCUGUGG---AGACAGCAAUGUUAGUAAUCAUA ...........(((((((((((((.....)))((..(((((((..(((((((.(((.((((((....)))))).))))))))))..))))))).---.))..))))))..))))...... ( -42.70) >consensus CUAUUGGCAGCUGCUCAAUGCGAUCUUCAAGCUCGGCGCAGGCAAGCAUCUUUGCAUUUAAGUCGCUGCUUAAUUGUAAAAUGUUUGCCUGUGG___AGAACAUAAUGUUAGUAAUCAUA .((((((((...((.....))...............(((((((((((((.((((((.((((((....)))))).))))))))))))))))))).............))))))))...... (-29.30 = -30.50 + 1.20)



| Location | 11,617,022 – 11,617,130 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -18.42 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11617022 108 + 27905053 GUCAAGCAUCUUUGCAUUUAAGUCGCUGCUUAAUUGUAAAAUGUUUGCCUGUGG---AGAACAUAAUGU---------UAUAAUCUAAUACAAGGAAAGCCAGUGUAACCCACCUCAAAC (.(((((((.((((((.((((((....)))))).))))))))))))).).((((---..(((.....))---------).........((((.((....))..))))..))))....... ( -23.40) >DroSec_CAF1 6958 120 + 1 GGUAAGCAACUUUGCAUUUAAGUCGCUGCUUAAUUGUAAAAUGCUUGCCUGUGGAGGAGAACAUAAUGUCAAUAAUCUUAUAACCUAAUACAAGGAAAGCCAGAGUAACACACCUCGCAA ((((((((..((((((.((((((....)))))).)))))).))))))))((((.(((.((........)).............(((......))).................))))))). ( -29.60) >DroSim_CAF1 7086 120 + 1 GGCAAGCAUCUUUGCAUUUAAGUCGCUGCUUAAUUGUAAAAUGUUUGCCUGUGGAGGAGAACAUAAUAUUAUUAAUCAUAUAACCUAAUACAAGGAAAUCCAUUGUAACCCACCUCGCAA (((((((((.((((((.((((((....)))))).)))))))))))))))((((.(((........((((........)))).......(((((((....)).))))).....))))))). ( -33.70) >DroEre_CAF1 4550 106 + 1 GGCAAGAAUCAUUGCAUUUAAGUCGGUGCUUAGUUGUAAGAUGUUUGCCUGUGG---AGAAAAUAAUGUUAGUAAUCCUAUAAGU-----------AAACCCGUGUAACCCACCUCGAAA ((((((.(((.(((((.((((((....)))))).)))))))).))))))....(---((..........(((.....))).....-----------......(((.....)))))).... ( -21.10) >DroYak_CAF1 5046 115 + 1 GGCUUGCAUCUUUGCAUUUAAGUCGGAGCUUAGUUGUAAGAUGUUUGCCUGUGG---AGACAGCAAUGUUAGUAAUCAUAUAAGUUGAUACAAGGAAAACC--AGUAACCCACCUCUUUU (((..(((((((.(((.((((((....)))))).))))))))))..))).((((---.((((....)))).(((.(((.......))))))..((....))--......))))....... ( -31.80) >consensus GGCAAGCAUCUUUGCAUUUAAGUCGCUGCUUAAUUGUAAAAUGUUUGCCUGUGG___AGAACAUAAUGUUAGUAAUCAUAUAACCUAAUACAAGGAAAACCAGUGUAACCCACCUCGAAA (((((((((.((((((.((((((....)))))).)))))))))))))))((((........))))....................................................... (-18.42 = -19.02 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:01 2006