
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,607,745 – 11,607,877 |
| Length | 132 |
| Max. P | 0.971941 |

| Location | 11,607,745 – 11,607,852 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 95.70 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -25.58 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11607745 107 + 27905053 ACGGAGCGUACGAAAAAUGCAAUUUUAAUUAGCCAGACGUGCGACAAACGCACGUAGAACGCAUAACCGCAAGAUGGUGCUGCAAUUAGGCGAACCCAUCCGAAUCC .((((((((.......))))...........(((..(((((((.....))))))).....(((..((((.....))))..))).....))).......))))..... ( -31.60) >DroSec_CAF1 137652 107 + 1 GCGAAGCGUACGAAAAAUGCAAUUUUAAUUAGCCAGACGUGCGACAAACGCACGUAGAACGCAUAACCGCAAGAUGGUGCUGCAAUUAGGCGAACCCAUCCGAAUCG (((..((((.......))))...........((...(((((((.....))))))).....)).....)))..(((((((((.......))))...)))))....... ( -27.60) >DroSim_CAF1 145801 107 + 1 ACGAAGCGUACGAAAAAUGCAAUUUUAAUUAGCCAGACGUGCGACAAACGCACGUAGAACGCAUAACCGCAAGAUGGUGCUGCAAUUAGGCGAACCCAUCCGAAUCG .(((.((((.......))))...........(((..(((((((.....))))))).....(((..((((.....))))..))).....))).............))) ( -27.20) >DroYak_CAF1 139131 107 + 1 ACGGAGCGUACGAAAAAUGCAAUUUUAAUUAGCCAGACGUGCGACAAACGCACGUAGAACGCAUAACCGCAAGAUGGUGCUGCAAUUAGGCGAAUCCAUACGAAUCG ..(((((((.......))))...........(((..(((((((.....))))))).....(((..((((.....))))..))).....)))...))).......... ( -29.60) >DroAna_CAF1 229061 107 + 1 GGGAAGCGUUCGAAAAAUGCAAUUUUAAUUAGCCAGACGUGCGACAAACGCACGUAGAACGCAUAACCGCAAGAUGGCGCUGCAAUUAGGCGAACCCAUCAGGAUCC .(((.((((((.......(((((....))).))...(((((((.....))))))).))))))....((....(((((((((.......))))...))))).)).))) ( -33.40) >consensus ACGAAGCGUACGAAAAAUGCAAUUUUAAUUAGCCAGACGUGCGACAAACGCACGUAGAACGCAUAACCGCAAGAUGGUGCUGCAAUUAGGCGAACCCAUCCGAAUCG .(((.((((.......))))...........(((..(((((((.....))))))).....(((..((((.....))))..))).....))).............))) (-25.58 = -25.50 + -0.08)

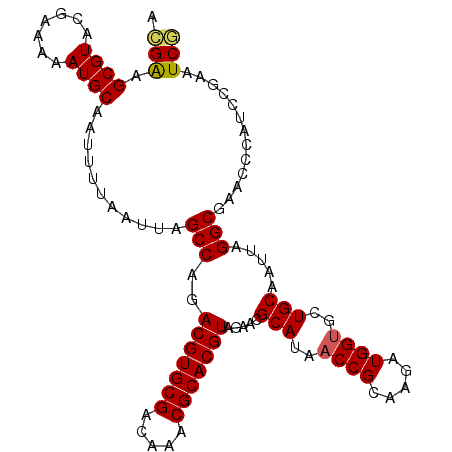
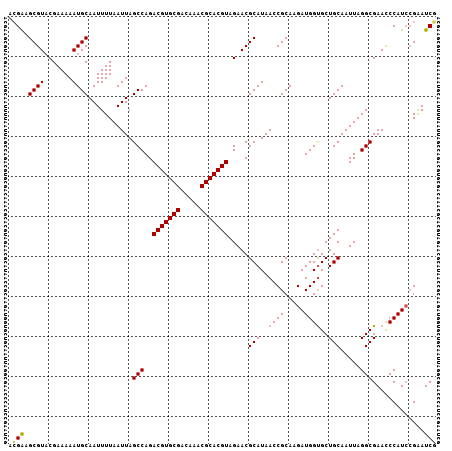
| Location | 11,607,745 – 11,607,852 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 95.70 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -26.24 |
| Energy contribution | -26.56 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11607745 107 - 27905053 GGAUUCGGAUGGGUUCGCCUAAUUGCAGCACCAUCUUGCGGUUAUGCGUUCUACGUGCGUUUGUCGCACGUCUGGCUAAUUAAAAUUGCAUUUUUCGUACGCUCCGU (((..((((((((((.((......))))).))))).(((((..(((((((..(((((((.....)))))))..))).(((....)))))))...))))))).))).. ( -32.80) >DroSec_CAF1 137652 107 - 1 CGAUUCGGAUGGGUUCGCCUAAUUGCAGCACCAUCUUGCGGUUAUGCGUUCUACGUGCGUUUGUCGCACGUCUGGCUAAUUAAAAUUGCAUUUUUCGUACGCUUCGC (((..((((((((((.((......))))).))))).(((((..(((((((..(((((((.....)))))))..))).(((....)))))))...)))))))..))). ( -32.80) >DroSim_CAF1 145801 107 - 1 CGAUUCGGAUGGGUUCGCCUAAUUGCAGCACCAUCUUGCGGUUAUGCGUUCUACGUGCGUUUGUCGCACGUCUGGCUAAUUAAAAUUGCAUUUUUCGUACGCUUCGU (((..((((((((((.((......))))).))))).(((((..(((((((..(((((((.....)))))))..))).(((....)))))))...)))))))..))). ( -33.00) >DroYak_CAF1 139131 107 - 1 CGAUUCGUAUGGAUUCGCCUAAUUGCAGCACCAUCUUGCGGUUAUGCGUUCUACGUGCGUUUGUCGCACGUCUGGCUAAUUAAAAUUGCAUUUUUCGUACGCUCCGU ((...((((((((...((.(((((((((.......))))))))).))(((..(((((((.....)))))))..))).................))))))))...)). ( -31.30) >DroAna_CAF1 229061 107 - 1 GGAUCCUGAUGGGUUCGCCUAAUUGCAGCGCCAUCUUGCGGUUAUGCGUUCUACGUGCGUUUGUCGCACGUCUGGCUAAUUAAAAUUGCAUUUUUCGAACGCUUCCC ((((((....))))))...(((((((((.......))))))))).((((((.(((((((.....)))))))...((...........)).......))))))..... ( -33.50) >consensus CGAUUCGGAUGGGUUCGCCUAAUUGCAGCACCAUCUUGCGGUUAUGCGUUCUACGUGCGUUUGUCGCACGUCUGGCUAAUUAAAAUUGCAUUUUUCGUACGCUUCGU ......(((((((((.((......))))).)))))).((((....((((.(.(((((((.....)))))))...((...........)).......).)))).)))) (-26.24 = -26.56 + 0.32)

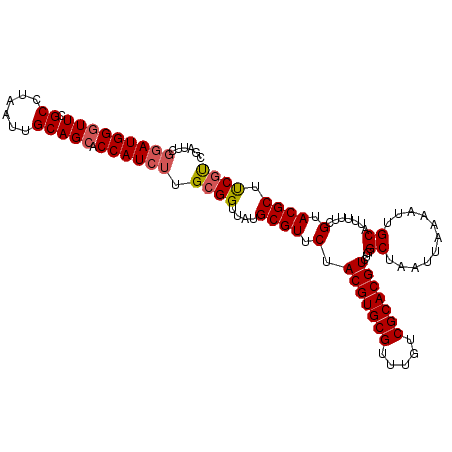

| Location | 11,607,777 – 11,607,877 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.82 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11607777 100 + 27905053 CCAGACGUGCGACAAACGCACGUAGAACGCAUAACCGCAAGAUGGUGCUGCAAUUAGGCG-----AACCCAUCCGAAUCCGAAUCAGAGUACGAAUCCGUAGCAG--------- ....(((((((.....))))))).............((..(((((((((.......))))-----...)))))................((((....))))))..--------- ( -23.80) >DroPse_CAF1 169276 97 + 1 GCAAACGUGCGACAAACGCACGUAGAAGGCAUAACCACAAGAUGGCGCUGCAAUUAGGCGAAACGAACCCAUCAGGAACGUUACCAGAGU--GACUCCA--------------- ((..(((((((.....))))))).....))..........(((((((((.......))))........))))).(((..(((((....))--)))))).--------------- ( -27.30) >DroSim_CAF1 145833 100 + 1 CCAGACGUGCGACAAACGCACGUAGAACGCAUAACCGCAAGAUGGUGCUGCAAUUAGGCG-----AACCCAUCCGAAUCGGAAUCAGAGUACGAAUCCGUAGCAG--------- (((.(((((((.....))))))).....((......))....)))((((((.....((..-----...)).........(((.((.......)).))))))))).--------- ( -27.60) >DroYak_CAF1 139163 96 + 1 CCAGACGUGCGACAAACGCACGUAGAACGCAUAACCGCAAGAUGGUGCUGCAAUUAGGCG-----AAUCCAUACGAAUCGGAAUCCGAAUCC-------------AUUCCGAGU ....(((((((.....))))))).....((((...(....)...))))(((......)))-----............(((((((........-------------))))))).. ( -26.00) >DroAna_CAF1 229093 98 + 1 CCAGACGUGCGACAAACGCACGUAGAACGCAUAACCGCAAGAUGGCGCUGCAAUUAGGCG-----AACCCAUCAGGAUCCUGACCC-CGUACC-GUCCAUACCCU--------- ....(((((((.....))))))).....((......))..(((((((((.......))))-----......((((....))))...-....))-)))........--------- ( -26.40) >DroPer_CAF1 170590 97 + 1 GCAAACGUGCGACAAACGCACGUAGAAGGCAUAACCACAAGAUGGCGCUGCAAUUAGGCGAAACGAACCCAUCAGGAACGUUACCAGAAU--GACUCCA--------------- ((..(((((((.....))))))).....))..........(((((((((.......))))........))))).(((..((((......)--)))))).--------------- ( -24.40) >consensus CCAGACGUGCGACAAACGCACGUAGAACGCAUAACCGCAAGAUGGCGCUGCAAUUAGGCG_____AACCCAUCAGAAUCGGAACCAGAGUACGAAUCCA_______________ ....(((((((.....))))))).................(((((((((.......))))........)))))......................................... (-17.90 = -17.82 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:56 2006