| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,657,208 – 1,657,320 |
| Length | 112 |
| Max. P | 0.546259 |

| Location | 1,657,208 – 1,657,320 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.88 |
| Mean single sequence MFE | -39.13 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.37 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1657208 112 + 27905053 AAUUAUAUCGGGCUGCGUCAUUCUGCCGCUGUCUCUGCCGCUGCUACUGCCUCUGCCACCGCGAUAUCAGGCU----UCUAGUAUCAG----CCUCUGUCGCUGUCGCCGACGCAGUGGG .........((((.(((.(.....).))).)))).........(((((((.((.((.((.((((((..(((((----.........))----))).)))))).)).)).)).))))))). ( -38.40) >DroSec_CAF1 4550 112 + 1 AAUUAUAUCGGACUGCGCCAUUCUGCCGCUGUCUCUGCCGCAGCCACUGCCUCUGCCACCGCGAUAUCAGGCU----UCUAGUAUCAG----CCUCUGUCGCAGUCGCCGACGCAGUGGG .........((......))...((((.((.......)).))))(((((((.((.((.((.((((((..(((((----.........))----))).)))))).)).)).)).))))))). ( -41.90) >DroSim_CAF1 5811 112 + 1 AAUUAUAUCGGACUGCGCCAUUCUGCCGCUGUCUCUGCCGCUGCCACUGCCUCUGCCACCGCGAUAUCAGGCU----UCUAGUAUCAG----CCUCUGUCGCAGUCGCCGACGCAGUGGG .........((((.(((.(.....).))).)))).........(((((((.((.((.((.((((((..(((((----.........))----))).)))))).)).)).)).))))))). ( -41.30) >DroEre_CAF1 6422 100 + 1 AAUUAUAUCGGGCUACGUAACUCAGCCG------------CGGACGCUGUCUCUGCCACCGCGAUAUCAGGCU----UCUAGUAUCAG----CCUCUGUCGCCGUCGCCGACGCAGCUGC .......(((((((.........)))).------------)))..(((((.((.((.((.((((((..(((((----.........))----))).)))))).)).)).)).)))))... ( -34.50) >DroYak_CAF1 6399 100 + 1 AAUUAUAUCGGGCGGCGCACUUCUGCCG------------CGGACGCUGCCUCUGCCACCGCGAUAUCAGGCU----UCUAGUAUCAG----CCUCUGUCGCAGUCGCCGACGCAGUUGC ...........((((((......)))))------------)....(((((.((.((.((.((((((..(((((----.........))----))).)))))).)).)).)).)))))... ( -37.20) >DroAna_CAF1 6837 114 + 1 AAUUAUGUCGGU------GGUUCGUGAGCUGCCUCGGCCAUUGCCUCGGCCUCUGCCACUGCUAUGUAGUGCUCUCGUCUGAUAUGGGGUUAGCUCUGUCGCAGUCGCAGUCGUCGUGGA ......(((((.------(((.(....)..)))))))).....((.((((..((((.(((((....(((.(((..(.((......)).)..))).)))..))))).))))..)))).)). ( -41.50) >consensus AAUUAUAUCGGGCUGCGCCAUUCUGCCGCUGUCUCUGCCGCUGCCACUGCCUCUGCCACCGCGAUAUCAGGCU____UCUAGUAUCAG____CCUCUGUCGCAGUCGCCGACGCAGUGGG ...........................................(((((((.((.((.((.((((((..(((.....................))).)))))).)).)).)).))))))). (-21.06 = -21.37 + 0.31)

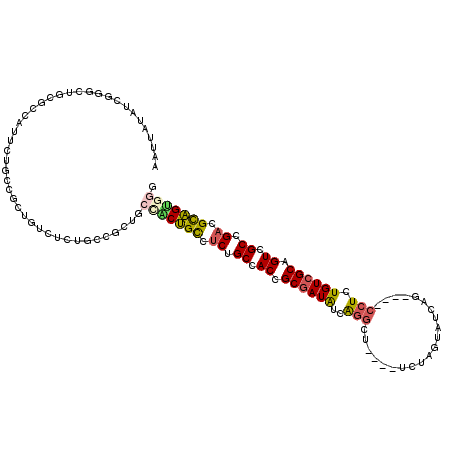
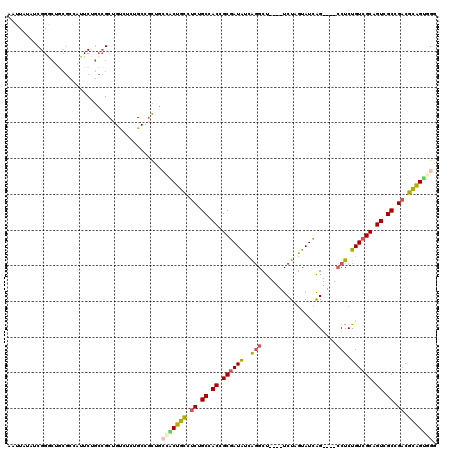
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:11 2006