
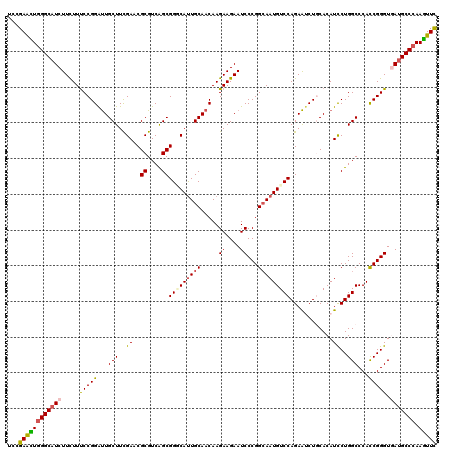
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,570,542 – 11,570,662 |
| Length | 120 |
| Max. P | 0.992709 |

| Location | 11,570,542 – 11,570,662 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -37.44 |
| Energy contribution | -37.33 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
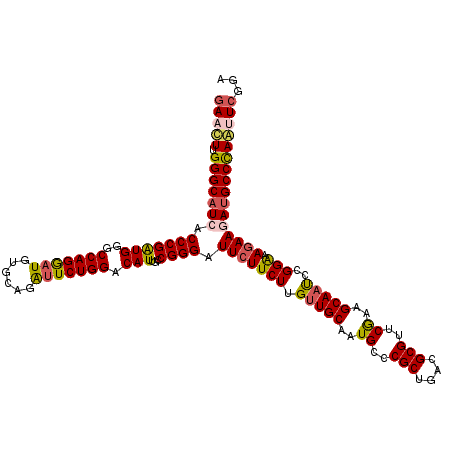
>3R_DroMel_CAF1 11570542 120 + 27905053 UCCGAAUUGGGCAUCUUCUUUCCGGAUUGCUUCGAACGCGUCAGCGGUCAUUGCAACAAGAAGAAUCCCGGCAAUGUCCAGAAUCUCCACAUCCUGGCCCAUCGGGUGAUGCCCAAAUUC ...((((((((((((((((((...(.((((...((.(((....))).))...)))))..)))))).(((((....(.((((............)))))...))))).))))))).))))) ( -38.90) >DroSec_CAF1 99712 120 + 1 UCCGAACUGGGCAUCUUCUUUCCGGAUUGCUUCGAACGCGUCAGCGGACAUUGCAACAAGAAGAAUCCCGGCAAUGUCCAGAAUCUGCACAUCCUGGCCCAUCGGGUUAUGCCCAAGUUC ...((((((((((..........((((((((((....((....))(((((((((.....((....))...))))))))).)))...))).))))((((((...))))))))))).))))) ( -43.40) >DroSim_CAF1 103573 120 + 1 UCUGAACUGGGCAUCUUCUUCCCGGAUUGCUUCGAACGCGUCAGCGGGCAUUGCAACAAGAAGAAUCCCGGCAAUGUCCAGAAUCUGCACAUCCUGGCCCAUCGGGUUAUGCCCAAGUUC ...((((((((((..........((((((((((....((....))(((((((((.....((....))...))))))))).)))...))).))))((((((...))))))))))).))))) ( -42.50) >DroEre_CAF1 102815 120 + 1 UCCGAAUUGGGCAUCUUCUUUCCGGAUUGCUUCGAACGCGUCAGCGGGCACUGCAACAAGAAGAAUCCCGGCAAUGUCCAGAAUCUGCACAUCCUGGCCCACCGGGUGAUGCCCAAAUUC ...((((((((((((((((((...(.((((...(..(((....)))..)...)))))..)))))).(((((....(.((((............)))))...))))).))))))).))))) ( -42.00) >DroYak_CAF1 100573 120 + 1 UCCGAGUUGGGCAUCUUCUUUCCGGAUUGCUUCGAACGCGUCGGCGGGCAUUGCAACAAGAAGAAUCCCGGCAAUGUCCAGAACCUGCACAUUCUGGCCCACCGGGUGAUGCCCAAGUUC ...((.(((((((((((((((...(.((((...(..(((....)))..)...)))))..)))))).(((((....(.((((((........)))))))...))))).))))))))).)). ( -45.10) >DroAna_CAF1 185701 120 + 1 AUAGAGCUAGGCUUCUUUUUUCCGGGCUGCUUUGAGCGCGUCAGCGGCCAUUGCACCAAGAAGAAUCCCGGAAAUGUCCAGAGCCUUCACAUCCUGGCCCACCGGGUGAUGCCCAAGUUU ...(((((((((((....((((((((...((((...(((....)))((....)).....))))...))))))))......))))))...((.(((((....))))))).......))))) ( -37.70) >consensus UCCGAACUGGGCAUCUUCUUUCCGGAUUGCUUCGAACGCGUCAGCGGGCAUUGCAACAAGAAGAAUCCCGGCAAUGUCCAGAAUCUGCACAUCCUGGCCCACCGGGUGAUGCCCAAGUUC ...((((((((((((.....(((((...(((..((..((....))(((((((((.....((....))...)))))))))............))..)))...))))).))))))).))))) (-37.44 = -37.33 + -0.11)


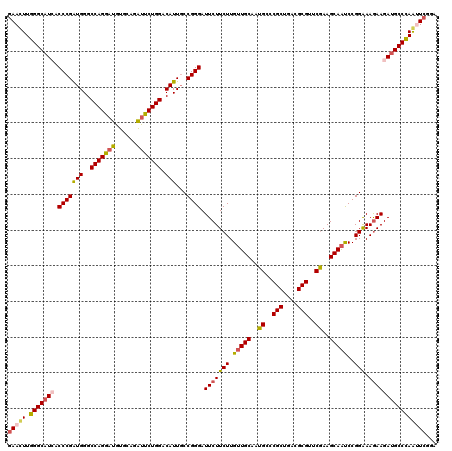
| Location | 11,570,542 – 11,570,662 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -49.53 |
| Consensus MFE | -42.75 |
| Energy contribution | -42.92 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11570542 120 - 27905053 GAAUUUGGGCAUCACCCGAUGGGCCAGGAUGUGGAGAUUCUGGACAUUGCCGGGAUUCUUCUUGUUGCAAUGACCGCUGACGCGUUCGAAGCAAUCCGGAAAGAAGAUGCCCAAUUCGGA (((((.(((((((.(((((((..(((((((......))))))).)))...)))).(((((((.(((((..(((.(((....))).)))..)))))..)).)))))))))))))))))... ( -48.60) >DroSec_CAF1 99712 120 - 1 GAACUUGGGCAUAACCCGAUGGGCCAGGAUGUGCAGAUUCUGGACAUUGCCGGGAUUCUUCUUGUUGCAAUGUCCGCUGACGCGUUCGAAGCAAUCCGGAAAGAAGAUGCCCAGUUCGGA (((((.((((((..((.(((..((..((((((((((....(((((((((((((((....)))))..))))))))))))).)))))))...)).))).)).......)))))))))))... ( -50.90) >DroSim_CAF1 103573 120 - 1 GAACUUGGGCAUAACCCGAUGGGCCAGGAUGUGCAGAUUCUGGACAUUGCCGGGAUUCUUCUUGUUGCAAUGCCCGCUGACGCGUUCGAAGCAAUCCGGGAAGAAGAUGCCCAGUUCAGA (((((.((((((..(((((((..(((((((......))))))).)))...)))).(((((((((((((..((..(((....)))..))..))))..))))))))).)))))))))))... ( -50.60) >DroEre_CAF1 102815 120 - 1 GAAUUUGGGCAUCACCCGGUGGGCCAGGAUGUGCAGAUUCUGGACAUUGCCGGGAUUCUUCUUGUUGCAGUGCCCGCUGACGCGUUCGAAGCAAUCCGGAAAGAAGAUGCCCAAUUCGGA (((((.(((((((.(((((..(.(((((((......)))))))...)..))))).(((((((.(((((..((..(((....)))..))..)))))..)).)))))))))))))))))... ( -55.40) >DroYak_CAF1 100573 120 - 1 GAACUUGGGCAUCACCCGGUGGGCCAGAAUGUGCAGGUUCUGGACAUUGCCGGGAUUCUUCUUGUUGCAAUGCCCGCCGACGCGUUCGAAGCAAUCCGGAAAGAAGAUGCCCAACUCGGA ((..(((((((((.(((((..(.(((((((......)))))))...)..))))).(((((((.(((((..((..(((....)))..))..)))))..)).)))))))))))))).))... ( -52.20) >DroAna_CAF1 185701 120 - 1 AAACUUGGGCAUCACCCGGUGGGCCAGGAUGUGAAGGCUCUGGACAUUUCCGGGAUUCUUCUUGGUGCAAUGGCCGCUGACGCGCUCAAAGCAGCCCGGAAAAAAGAAGCCUAGCUCUAU ......((((.((((((((....)).))..))))(((((((.....((((((((..((.....))(((..((..(((....)))..))..))).))))))))..)).))))).))))... ( -39.50) >consensus GAACUUGGGCAUCACCCGAUGGGCCAGGAUGUGCAGAUUCUGGACAUUGCCGGGAUUCUUCUUGUUGCAAUGCCCGCUGACGCGUUCGAAGCAAUCCGGAAAGAAGAUGCCCAAUUCGGA (((((.(((((((.(((((((..(((((((......))))))).)))...)))).(((((((.(((((..((..(((....)))..))..)))))..))).))))))))))))))))... (-42.75 = -42.92 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:39 2006