| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,566,600 – 11,566,711 |
| Length | 111 |
| Max. P | 0.562578 |

| Location | 11,566,600 – 11,566,711 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.55 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -13.18 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11566600 111 - 27905053 GUGGACAGCGGAGGGGGGGGGGGAGUGGGCUGAAAAGGACAGGCAGAAUCGACCCAUAAA-AAACGAGAGCGAAUAAAAAGCGCAAGAAAAUUACUUUCCGCUCUACAAAUU (((((..((((((((..(((..((...(.(((.......))).)....))..))).....-........(((.........)))..........)))))))))))))..... ( -32.50) >DroPse_CAF1 94636 93 - 1 GUGUGCA-------------GAGAGUGGAGUGG-----ACAGG-AAAAUCGACCCAUAAAAAAACGAGAGCAAAUAAAAAGCGCAAGAAAAUUACUUUCCGCUCUACAAAAU .......-------------....(((((((((-----(.((.-....(((.............)))..............(....).......)).))))))))))..... ( -19.52) >DroSec_CAF1 95729 105 - 1 GUGUACAACA------GGAGGAGAGUGGACUGAAAAGGACAGGCAGAGUCGACCCAUAAA-AAACGAGAGCGAAUAAAAAGCGCAAGAAAAUUACUUUCCGCUCUACAAAUU ..........------...(.((((((((..(....((...(((...)))...)).....-........(((.........)))..........)..)))))))).)..... ( -18.30) >DroYak_CAF1 96418 104 - 1 GUGGACAACA------G-AGAGGAGUGGACUGUAAAGGACAGGCAGAAUCGACCCAUAAA-AAACGAGAGCGAAUAAAAAGCGCAAGAAAAUUACUUUCCGCUCUACAAAUU ..........------.-.(.((((((((..((((.......((....(((.........-...)))..((.........)))).......))))..)))))))).)..... ( -19.74) >DroAna_CAF1 180600 96 - 1 GCG---------------AUGAGAGGGGAGGGUAGAGCGAUGGCAAAAUCGACCCAUAAA-AAACGAGAGCGAAUAAAAAGCGCAAGAAAAUUACUUUCCGCUCUACAAAUU ...---------------.((((((.(((((((((..((((......)))).........-........(((.........))).......))))))))).)))).)).... ( -24.40) >DroPer_CAF1 95924 93 - 1 GUGUGCA-------------GAGAGUGGAGUGG-----ACAGG-AAAAUCGACCCAUAAAAAAACGAGAGCAAAUAAAAAGCGCAAGAAAAUUACUUUCCGCUCUACAAAAU .......-------------....(((((((((-----(.((.-....(((.............)))..............(....).......)).))))))))))..... ( -19.52) >consensus GUGUACA___________AGGAGAGUGGACUGAAAAGGACAGGCAAAAUCGACCCAUAAA_AAACGAGAGCGAAUAAAAAGCGCAAGAAAAUUACUUUCCGCUCUACAAAUU .....................((((((((..(................(((.............)))..(((.........)))..........)..))))))))....... (-13.18 = -13.32 + 0.14)

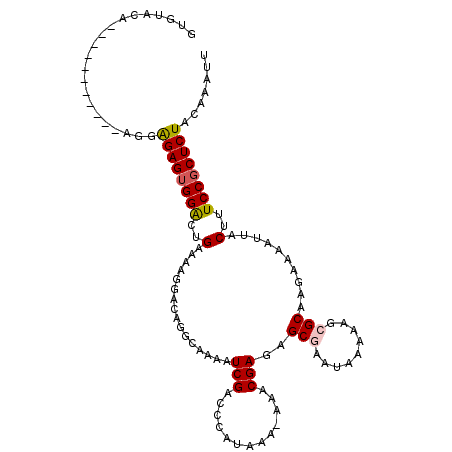

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:37 2006