| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,530,453 – 11,530,547 |
| Length | 94 |
| Max. P | 0.831227 |

| Location | 11,530,453 – 11,530,547 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -27.68 |
| Energy contribution | -28.38 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
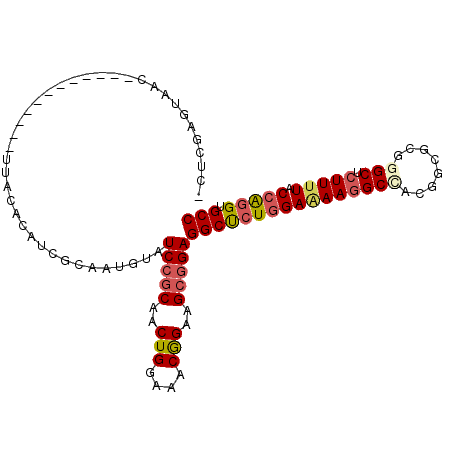
>3R_DroMel_CAF1 11530453 94 + 27905053 -CUCGAGUAAC------------UUCCACAUCGCAAUGUAUCCGCAACUGGAAACAGAAGCGGAGGCUCUGGAAAAGGCUACGGCACGGGCUCUUUUACCAGGUGCC -..........------------....((((....)))).(((((..(((....)))..)))))((((((((((((((((........))).))))).))))).))) ( -33.70) >DroSec_CAF1 64449 94 + 1 -CUCGAGUAAC------------UUCCACAUCGCAAUGUAUCCGCAACUGGAAACGGAAGCGGAGGCUCUGGAGAAGGCUACGGCGCGGGCUCUUUUACCAGGUGCC -.....(((.(------------(((..((..((......(((((..(((....)))..))))).))..))..))))..)))(((((.((........))..))))) ( -33.80) >DroSim_CAF1 66578 94 + 1 -CCCGAGUAAC------------UUCCACAUCGCAAUGUAUCCGCAACUGGAAACGGAAGCGGAGGCUCUGGAAAAGGCAACGGCGCGGGCUCUUUUACCAGGUGCC -((.((((...------------....((((....)))).(((((..(((....)))..))))).)))).))....(....)(((((.((........))..))))) ( -32.50) >DroEre_CAF1 67424 94 + 1 -UUCGAGUAUC------------UCACGCUGCGCAAUGUAUCCGCAACUGGAAACGGAAGCGGAGGCUCUGGAAAAGGCCACGGCGCGAGCCCUUUUACCAGGUGCC -...(((...)------------)).....((((...((.(((((..(((....)))..))))).)).((((.(((((....(((....)))))))).)))))))). ( -36.80) >DroYak_CAF1 62907 95 + 1 ACUCGAGUAAC------------UUACUCUGCGCCAUGUAUCCGCAACUGGAAACGGAAGCGGAGGCUCUGGAAAAGGCCACGGCGCGGGCUCUUUUACCAGGUGCC ....(((((..------------.)))))((((((.....(((((..(((....)))..)))))(((..(....)..)))..))))))(((.(((.....))).))) ( -40.80) >DroAna_CAF1 150572 103 + 1 -ACCAGGUAUCACGGAAGUCAACUAAUAUA---UGAUGUAUCCCCAACUGGAAACGGAAGCUGAGGCCAUUGAAAACGCCACAGCCCGGGCCCUUCUACCGGGUGCC -....((((((.(((..((((.........---)))).....(((..(((....)))..((((.(((..........))).))))..)))........))))))))) ( -31.30) >consensus _CUCGAGUAAC____________UUACACAUCGCAAUGUAUCCGCAACUGGAAACGGAAGCGGAGGCUCUGGAAAAGGCCACGGCGCGGGCUCUUUUACCAGGUGCC ........................................(((((..(((....)))..)))))((((((((((((((((........))).))))).))))).))) (-27.68 = -28.38 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:29 2006