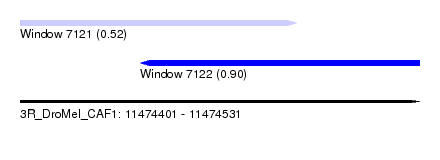
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,474,401 – 11,474,531 |
| Length | 130 |
| Max. P | 0.900954 |

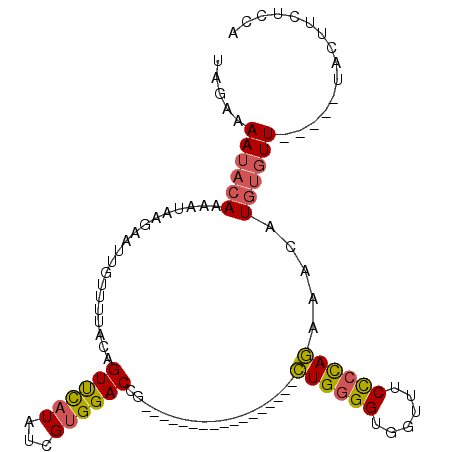
| Location | 11,474,401 – 11,474,491 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -13.20 |
| Energy contribution | -14.15 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
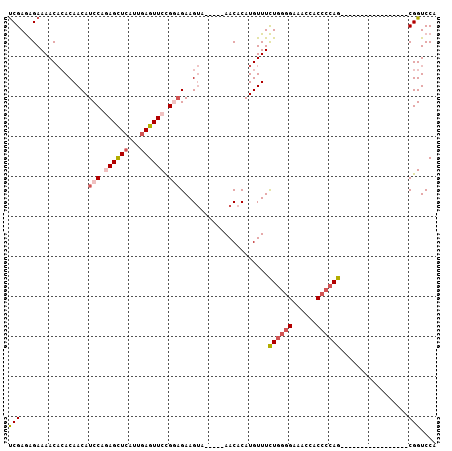
>3R_DroMel_CAF1 11474401 90 + 27905053 UCGAGAGAAAACACACAACAUCCAGAGCUCAUUGAGUUCUGGAGAAGUAAACUAAACACAUGUUUCUGGGGAAACCACCCCAG-----------------CGGUCCA ((....))(((((.......((((((((((...))))))))))..((....)).......)))))((((((......))))))-----------------....... ( -27.00) >DroSec_CAF1 8590 85 + 1 UCGAGAGAAAACACACAACAUCCAGAGCUCAUUGAGUUCUGGAGAAGUA-----AACACAUGUUUCUGGGGAAACCACCCCAG-----------------CGGUCCA ((....))(((((.......((((((((((...))))))))))...((.-----...)).)))))((((((......))))))-----------------....... ( -26.90) >DroSim_CAF1 8815 85 + 1 UCGAGAGAAAACACACAACAUCCACAGCUCAUUGAGUUCCGGAGAAGUA-----AACACAUGUUUCUGGGGAAACCACCCCAG-----------------CGGUCCA ((....))(((((.......(((..(((((...)))))..)))...((.-----...)).)))))((((((......))))))-----------------....... ( -17.90) >DroEre_CAF1 10913 97 + 1 UCGAGAGAAAACACACAACAUCCAGAGCUCAUUGAGUUCCGAAGAAGUA-----AACACAUGUUUCUGGGGAAUCCACCCCAGCAGAGGAAC-----UUGCGGUCCA ((....))................(..((...((((((((........(-----(((....))))((((((......))))))....)))))-----))).))..). ( -22.70) >DroYak_CAF1 5998 102 + 1 UCGAGAGAAAACACACAACAUCCAGAGCUCAUUGAGUUCUGGCGAAGUA-----AACACAUGUUUCUGGGGAAUCCACCCCAGCAGAGGACCACGAUAUGCGGUCCG ((....))(((((........(((((((((...)))))))))....((.-----...)).)))))((((((......))))))....(((((.(.....).))))). ( -33.40) >DroAna_CAF1 96835 89 + 1 CCGAAGGAAAACCGACAACAUCCAUAGUUCGUUUAGUUCCG-AGAAGCG-----AACACAUGUUUUUAUAGAUCCAACCCCAACA------------AUGCGGUCCA .....(((...(((.((.........((((((((.......-..)))))-----)))...((..((....))..)).........------------.)))))))). ( -14.50) >consensus UCGAGAGAAAACACACAACAUCCAGAGCUCAUUGAGUUCCGGAGAAGUA_____AACACAUGUUUCUGGGGAAACCACCCCAG_________________CGGUCCA (((.................(((.((((((...)))))).)))......................((((((......)))))).................))).... (-13.20 = -14.15 + 0.95)

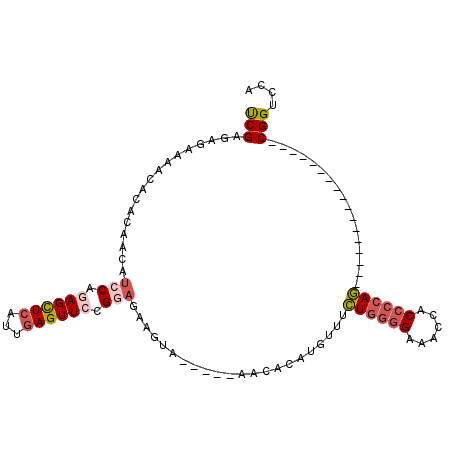
| Location | 11,474,440 – 11,474,531 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.65 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -15.26 |
| Energy contribution | -15.18 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11474440 91 - 27905053 UAGAAAAUACAAAAUAAGAAUUGUUUUACAGUUCAUAUCGUGGACCG-----------------CUGGGGUGGUUUCCCCAGAAACAUGUGUUUAGUUUACUUCUCCA .((((............((((((.....)))))).....((((((..-----------------((((((......))))))((((....)))).))))))))))... ( -24.00) >DroSec_CAF1 8629 85 - 1 UAGAAAAUACAAAAUAAGAAUUGUUUUU-UGUUCAUAUCGUGGACCG-----------------CUGGGGUGGUUUCCCCAGAAACAUGUGUU-----UACUUCUCCA .((.(((((((.................-.((((((...))))))..-----------------((((((......)))))).....))))))-----).))...... ( -19.90) >DroSim_CAF1 8854 86 - 1 UAGAAAAUACAAAAUAAGAAUUGUUUUACAGUUCAUAUCGUGGACCG-----------------CUGGGGUGGUUUCCCCAGAAACAUGUGUU-----UACUUCUCCG .((.((((((.......((((((.....)))))).....(((.....-----------------((((((......))))))...))))))))-----).))...... ( -23.00) >DroEre_CAF1 10952 98 - 1 CGGAAAAUACAAAAUAAGAAUUGUUUUACAGUCCAUUUCGUGGACCGCAA-----GUUCCUCUGCUGGGGUGGAUUCCCCAGAAACAUGUGUU-----UACUUCUUCG .((.(((((((.....(((...........((((((...))))))(....-----)....))).((((((......)))))).....))))))-----).))...... ( -24.40) >DroYak_CAF1 6037 103 - 1 UAGAAAAUACAAAAUAAGAACUGUUUUGCAGUCCAUAUCGCGGACCGCAUAUCGUGGUCCUCUGCUGGGGUGGAUUCCCCAGAAACAUGUGUU-----UACUUCGCCA .((.(((((((.....(((((((.....)))).........(((((((.....)))))))))).((((((......)))))).....))))))-----).))...... ( -33.00) >DroAna_CAF1 96874 84 - 1 CUGAAAAGUGAAAAUCGGGACUG------AGUUUAUAUCGUGGACCGCAU------------UGUUGGGGUUGGAUCUAUAAAAACAUGUGUU-----CGCUUCU-CG ..((.(((((((...((....((------..((((((...(.((((.((.------------...)).)))).)...))))))..))))..))-----))))).)-). ( -19.10) >consensus UAGAAAAUACAAAAUAAGAAUUGUUUUACAGUUCAUAUCGUGGACCG_________________CUGGGGUGGUUUCCCCAGAAACAUGUGUU_____UACUUCUCCA .....((((((...................((((((...))))))...................((((((......)))))).....))))))............... (-15.26 = -15.18 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:08 2006